
Oscillibacter sp. KLE 1745
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Oscillospiraceae; Oscillibacter; unclassified Oscillibacter
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
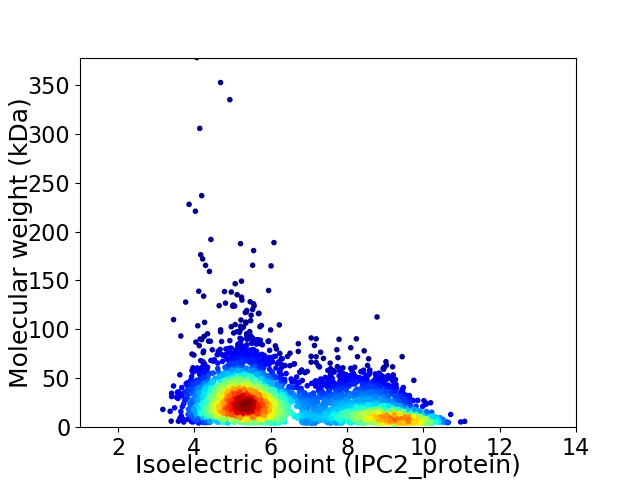
Virtual 2D-PAGE plot for 4287 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U2QRF8|U2QRF8_9FIRM AAA domain-containing protein OS=Oscillibacter sp. KLE 1745 OX=1226323 GN=HMPREF1546_03487 PE=4 SV=1
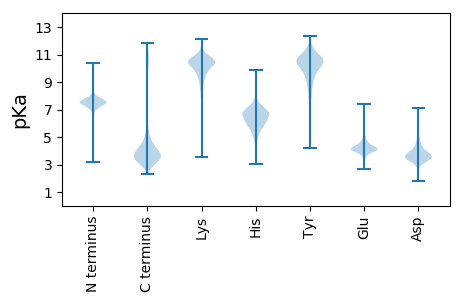
MM1 pKa = 7.57KK2 pKa = 10.55KK3 pKa = 9.03MLSMLLALILVFSLVGCSGSSGDD26 pKa = 5.07SGTADD31 pKa = 2.96TGGDD35 pKa = 3.16SSAAEE40 pKa = 3.97EE41 pKa = 4.25SDD43 pKa = 3.82VIKK46 pKa = 10.67IGLYY50 pKa = 9.7TPLTGTSALVGTQEE64 pKa = 4.58QMGVDD69 pKa = 4.06LAVKK73 pKa = 9.69QLNEE77 pKa = 3.92AGGVNGKK84 pKa = 8.56QFSVIAYY91 pKa = 8.63DD92 pKa = 3.61DD93 pKa = 3.48QFNAEE98 pKa = 4.18TAVKK102 pKa = 10.28VVTRR106 pKa = 11.84LTQTDD111 pKa = 3.62NVDD114 pKa = 4.05AIIGSMSSANILATADD130 pKa = 3.25IVEE133 pKa = 4.19QAQVLEE139 pKa = 4.64IGCGTSPTWTNAGYY153 pKa = 10.34EE154 pKa = 4.17YY155 pKa = 11.04VFRR158 pKa = 11.84GTQNAASFNTGIVEE172 pKa = 4.19LMTRR176 pKa = 11.84MGVTRR181 pKa = 11.84LGTMVSSTEE190 pKa = 3.91YY191 pKa = 10.02ATTGWAEE198 pKa = 4.25VKK200 pKa = 10.23EE201 pKa = 4.11QLADD205 pKa = 3.41TDD207 pKa = 3.81IEE209 pKa = 4.5IVLEE213 pKa = 4.06TDD215 pKa = 3.45YY216 pKa = 10.94MAGDD220 pKa = 3.76TDD222 pKa = 3.94FTGQITRR229 pKa = 11.84LLNADD234 pKa = 3.56LDD236 pKa = 4.82GILLYY241 pKa = 10.77GATEE245 pKa = 4.69DD246 pKa = 4.43YY247 pKa = 11.18GIEE250 pKa = 4.2CKK252 pKa = 8.43QLRR255 pKa = 11.84QLGYY259 pKa = 8.95EE260 pKa = 4.31GYY262 pKa = 9.75IYY264 pKa = 10.74GSEE267 pKa = 4.05TFAATDD273 pKa = 3.48VRR275 pKa = 11.84EE276 pKa = 4.34VASDD280 pKa = 3.39AANGVLFACGYY291 pKa = 8.18VIPDD295 pKa = 4.88AIEE298 pKa = 4.18DD299 pKa = 3.69AATEE303 pKa = 4.11EE304 pKa = 4.2EE305 pKa = 4.67KK306 pKa = 11.19AFLEE310 pKa = 4.44AFVEE314 pKa = 5.26EE315 pKa = 4.59YY316 pKa = 11.54GEE318 pKa = 4.31MPVSDD323 pKa = 3.65TAYY326 pKa = 10.22RR327 pKa = 11.84GYY329 pKa = 11.28DD330 pKa = 3.42SMMLLAKK337 pKa = 10.39VFEE340 pKa = 4.62TAEE343 pKa = 4.19SMEE346 pKa = 4.4GPDD349 pKa = 5.03LRR351 pKa = 11.84EE352 pKa = 3.73ALLNVDD358 pKa = 3.41YY359 pKa = 10.83TGIGGKK365 pKa = 9.45FDD367 pKa = 3.92YY368 pKa = 10.94SDD370 pKa = 3.73GSGDD374 pKa = 3.66GLEE377 pKa = 4.4GCNLYY382 pKa = 10.94AIVDD386 pKa = 4.16GKK388 pKa = 10.37NVPFEE393 pKa = 4.19TFLADD398 pKa = 5.26LNNQQ402 pKa = 3.21
MM1 pKa = 7.57KK2 pKa = 10.55KK3 pKa = 9.03MLSMLLALILVFSLVGCSGSSGDD26 pKa = 5.07SGTADD31 pKa = 2.96TGGDD35 pKa = 3.16SSAAEE40 pKa = 3.97EE41 pKa = 4.25SDD43 pKa = 3.82VIKK46 pKa = 10.67IGLYY50 pKa = 9.7TPLTGTSALVGTQEE64 pKa = 4.58QMGVDD69 pKa = 4.06LAVKK73 pKa = 9.69QLNEE77 pKa = 3.92AGGVNGKK84 pKa = 8.56QFSVIAYY91 pKa = 8.63DD92 pKa = 3.61DD93 pKa = 3.48QFNAEE98 pKa = 4.18TAVKK102 pKa = 10.28VVTRR106 pKa = 11.84LTQTDD111 pKa = 3.62NVDD114 pKa = 4.05AIIGSMSSANILATADD130 pKa = 3.25IVEE133 pKa = 4.19QAQVLEE139 pKa = 4.64IGCGTSPTWTNAGYY153 pKa = 10.34EE154 pKa = 4.17YY155 pKa = 11.04VFRR158 pKa = 11.84GTQNAASFNTGIVEE172 pKa = 4.19LMTRR176 pKa = 11.84MGVTRR181 pKa = 11.84LGTMVSSTEE190 pKa = 3.91YY191 pKa = 10.02ATTGWAEE198 pKa = 4.25VKK200 pKa = 10.23EE201 pKa = 4.11QLADD205 pKa = 3.41TDD207 pKa = 3.81IEE209 pKa = 4.5IVLEE213 pKa = 4.06TDD215 pKa = 3.45YY216 pKa = 10.94MAGDD220 pKa = 3.76TDD222 pKa = 3.94FTGQITRR229 pKa = 11.84LLNADD234 pKa = 3.56LDD236 pKa = 4.82GILLYY241 pKa = 10.77GATEE245 pKa = 4.69DD246 pKa = 4.43YY247 pKa = 11.18GIEE250 pKa = 4.2CKK252 pKa = 8.43QLRR255 pKa = 11.84QLGYY259 pKa = 8.95EE260 pKa = 4.31GYY262 pKa = 9.75IYY264 pKa = 10.74GSEE267 pKa = 4.05TFAATDD273 pKa = 3.48VRR275 pKa = 11.84EE276 pKa = 4.34VASDD280 pKa = 3.39AANGVLFACGYY291 pKa = 8.18VIPDD295 pKa = 4.88AIEE298 pKa = 4.18DD299 pKa = 3.69AATEE303 pKa = 4.11EE304 pKa = 4.2EE305 pKa = 4.67KK306 pKa = 11.19AFLEE310 pKa = 4.44AFVEE314 pKa = 5.26EE315 pKa = 4.59YY316 pKa = 11.54GEE318 pKa = 4.31MPVSDD323 pKa = 3.65TAYY326 pKa = 10.22RR327 pKa = 11.84GYY329 pKa = 11.28DD330 pKa = 3.42SMMLLAKK337 pKa = 10.39VFEE340 pKa = 4.62TAEE343 pKa = 4.19SMEE346 pKa = 4.4GPDD349 pKa = 5.03LRR351 pKa = 11.84EE352 pKa = 3.73ALLNVDD358 pKa = 3.41YY359 pKa = 10.83TGIGGKK365 pKa = 9.45FDD367 pKa = 3.92YY368 pKa = 10.94SDD370 pKa = 3.73GSGDD374 pKa = 3.66GLEE377 pKa = 4.4GCNLYY382 pKa = 10.94AIVDD386 pKa = 4.16GKK388 pKa = 10.37NVPFEE393 pKa = 4.19TFLADD398 pKa = 5.26LNNQQ402 pKa = 3.21
Molecular weight: 42.86 kDa
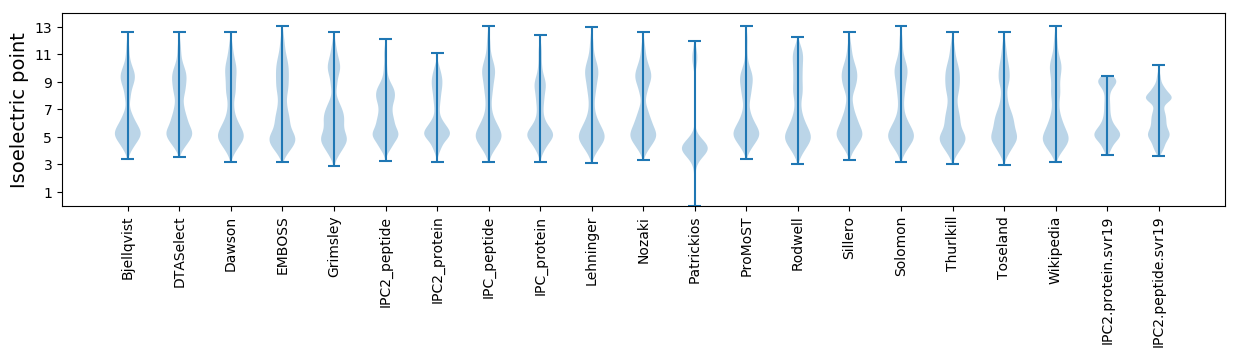
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U2R0V6|U2R0V6_9FIRM Putative anaerobic ribonucleoside-triphosphate reductase activating protein (Fragment) OS=Oscillibacter sp. KLE 1745 OX=1226323 GN=HMPREF1546_02791 PE=4 SV=1
MM1 pKa = 7.73FFRR4 pKa = 11.84HH5 pKa = 4.4WRR7 pKa = 11.84KK8 pKa = 9.3NRR10 pKa = 11.84GSRR13 pKa = 11.84RR14 pKa = 11.84FPLAPLSRR22 pKa = 11.84MMRR25 pKa = 11.84GRR27 pKa = 11.84WKK29 pKa = 10.12YY30 pKa = 7.26QWRR33 pKa = 11.84LLPLFFRR40 pKa = 11.84MKK42 pKa = 10.58VSLNN46 pKa = 3.41
MM1 pKa = 7.73FFRR4 pKa = 11.84HH5 pKa = 4.4WRR7 pKa = 11.84KK8 pKa = 9.3NRR10 pKa = 11.84GSRR13 pKa = 11.84RR14 pKa = 11.84FPLAPLSRR22 pKa = 11.84MMRR25 pKa = 11.84GRR27 pKa = 11.84WKK29 pKa = 10.12YY30 pKa = 7.26QWRR33 pKa = 11.84LLPLFFRR40 pKa = 11.84MKK42 pKa = 10.58VSLNN46 pKa = 3.41
Molecular weight: 5.96 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1076406 |
26 |
3491 |
251.1 |
27.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.886 ± 0.063 | 1.689 ± 0.019 |
5.579 ± 0.043 | 6.813 ± 0.038 |
3.818 ± 0.028 | 7.957 ± 0.038 |
1.84 ± 0.02 | 5.44 ± 0.037 |
4.529 ± 0.034 | 9.904 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.735 ± 0.02 | 3.128 ± 0.028 |
4.305 ± 0.024 | 3.569 ± 0.024 |
6.116 ± 0.05 | 5.432 ± 0.034 |
5.682 ± 0.046 | 7.016 ± 0.037 |
1.166 ± 0.016 | 3.397 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
