
Suttonella ornithocola
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Cardiobacteriales; Cardiobacteriaceae; Suttonella
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
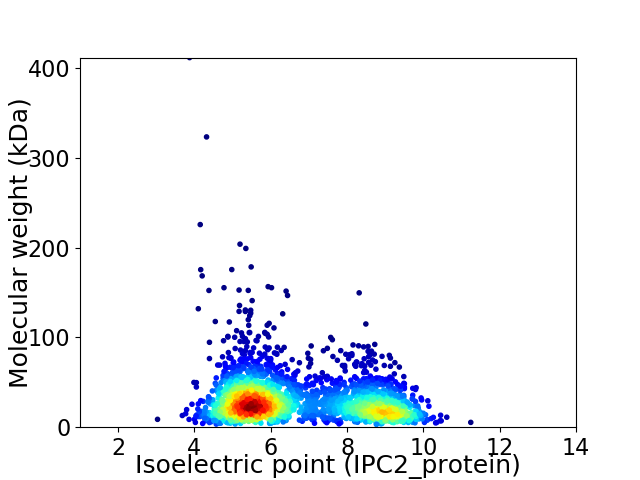
Virtual 2D-PAGE plot for 2483 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A380MQZ8|A0A380MQZ8_9GAMM 3-oxoacyl-[acyl-carrier-protein] reductase FabG OS=Suttonella ornithocola OX=279832 GN=fabG_1 PE=3 SV=1
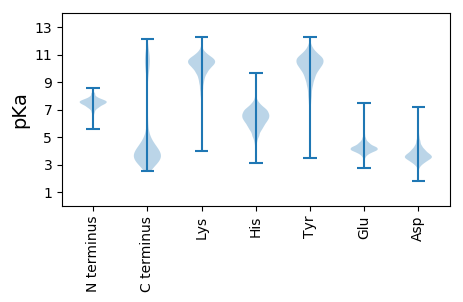
MM1 pKa = 7.08NHH3 pKa = 5.75STLQQLIEE11 pKa = 4.37ALEE14 pKa = 4.17EE15 pKa = 4.86DD16 pKa = 4.72DD17 pKa = 5.55FQFNPPATEE26 pKa = 4.35AEE28 pKa = 4.14LSAFEE33 pKa = 4.29AQHH36 pKa = 5.96QIEE39 pKa = 4.82LSPLFKK45 pKa = 10.73ALYY48 pKa = 9.54LKK50 pKa = 11.09ANGSVSTQTVDD61 pKa = 2.88MDD63 pKa = 3.97ALSDD67 pKa = 3.67FMEE70 pKa = 4.97PSEE73 pKa = 4.79HH74 pKa = 6.45GLSKK78 pKa = 11.01EE79 pKa = 3.99DD80 pKa = 3.8LQNMSLTDD88 pKa = 3.1RR89 pKa = 11.84WLSLTEE95 pKa = 4.14IEE97 pKa = 4.51TAIQTICQKK106 pKa = 11.1AEE108 pKa = 4.04DD109 pKa = 4.29YY110 pKa = 10.96FGNDD114 pKa = 2.77WQDD117 pKa = 3.26IILEE121 pKa = 3.91QDD123 pKa = 3.73DD124 pKa = 3.6NHH126 pKa = 7.95GEE128 pKa = 4.03YY129 pKa = 10.3KK130 pKa = 10.39PYY132 pKa = 10.17PFHH135 pKa = 6.57TRR137 pKa = 11.84WLPISKK143 pKa = 10.34NSLEE147 pKa = 4.47QYY149 pKa = 10.24FCLDD153 pKa = 4.21FDD155 pKa = 4.05PQEE158 pKa = 4.69DD159 pKa = 4.56GEE161 pKa = 4.39IGQVLLINIGDD172 pKa = 3.8EE173 pKa = 4.78SIDD176 pKa = 3.79EE177 pKa = 4.19DD178 pKa = 3.79WEE180 pKa = 4.55VYY182 pKa = 10.52RR183 pKa = 11.84MANSFEE189 pKa = 4.13NWVEE193 pKa = 4.31SYY195 pKa = 10.0WLGEE199 pKa = 4.17EE200 pKa = 4.32EE201 pKa = 5.24SDD203 pKa = 3.97IATLFDD209 pKa = 5.02EE210 pKa = 5.51IGQEE214 pKa = 4.06MGVLTSDD221 pKa = 4.35GEE223 pKa = 4.52FNQEE227 pKa = 3.6GYY229 pKa = 10.28FRR231 pKa = 11.84SAVHH235 pKa = 5.44EE236 pKa = 4.03HH237 pKa = 6.44LMNQFGDD244 pKa = 3.92EE245 pKa = 3.98NFMVIDD251 pKa = 3.64EE252 pKa = 4.53TEE254 pKa = 4.17YY255 pKa = 10.78PGDD258 pKa = 3.93ALYY261 pKa = 10.58DD262 pKa = 3.72IYY264 pKa = 10.84WFAPEE269 pKa = 3.91EE270 pKa = 3.88QRR272 pKa = 11.84PYY274 pKa = 10.6HH275 pKa = 6.65ALATNGLSTIEE286 pKa = 4.23MPSAEE291 pKa = 5.56DD292 pKa = 3.88GDD294 pKa = 4.12TKK296 pKa = 10.92NTFEE300 pKa = 5.47HH301 pKa = 6.79IEE303 pKa = 4.02LLAFFPPEE311 pKa = 3.72YY312 pKa = 10.09FLNGDD317 pKa = 4.0QIQMTEE323 pKa = 3.88QQSWILAVFKK333 pKa = 10.25TLAEE337 pKa = 4.04LLKK340 pKa = 9.96EE341 pKa = 4.18GQRR344 pKa = 11.84IDD346 pKa = 3.3INSTFEE352 pKa = 3.99LSGVEE357 pKa = 3.78ALPFKK362 pKa = 10.27EE363 pKa = 3.44ISLRR367 pKa = 11.84YY368 pKa = 8.25STLFTAQEE376 pKa = 4.21TLISTAHH383 pKa = 6.37GEE385 pKa = 4.4SIRR388 pKa = 11.84QLVIVPLYY396 pKa = 10.52SAEE399 pKa = 3.78QQYY402 pKa = 10.48QYY404 pKa = 10.96EE405 pKa = 4.26YY406 pKa = 11.13GYY408 pKa = 11.35AALNQTLKK416 pKa = 10.61AANVGDD422 pKa = 4.31IINPQRR428 pKa = 11.84ANAVSQQQ435 pKa = 3.05
MM1 pKa = 7.08NHH3 pKa = 5.75STLQQLIEE11 pKa = 4.37ALEE14 pKa = 4.17EE15 pKa = 4.86DD16 pKa = 4.72DD17 pKa = 5.55FQFNPPATEE26 pKa = 4.35AEE28 pKa = 4.14LSAFEE33 pKa = 4.29AQHH36 pKa = 5.96QIEE39 pKa = 4.82LSPLFKK45 pKa = 10.73ALYY48 pKa = 9.54LKK50 pKa = 11.09ANGSVSTQTVDD61 pKa = 2.88MDD63 pKa = 3.97ALSDD67 pKa = 3.67FMEE70 pKa = 4.97PSEE73 pKa = 4.79HH74 pKa = 6.45GLSKK78 pKa = 11.01EE79 pKa = 3.99DD80 pKa = 3.8LQNMSLTDD88 pKa = 3.1RR89 pKa = 11.84WLSLTEE95 pKa = 4.14IEE97 pKa = 4.51TAIQTICQKK106 pKa = 11.1AEE108 pKa = 4.04DD109 pKa = 4.29YY110 pKa = 10.96FGNDD114 pKa = 2.77WQDD117 pKa = 3.26IILEE121 pKa = 3.91QDD123 pKa = 3.73DD124 pKa = 3.6NHH126 pKa = 7.95GEE128 pKa = 4.03YY129 pKa = 10.3KK130 pKa = 10.39PYY132 pKa = 10.17PFHH135 pKa = 6.57TRR137 pKa = 11.84WLPISKK143 pKa = 10.34NSLEE147 pKa = 4.47QYY149 pKa = 10.24FCLDD153 pKa = 4.21FDD155 pKa = 4.05PQEE158 pKa = 4.69DD159 pKa = 4.56GEE161 pKa = 4.39IGQVLLINIGDD172 pKa = 3.8EE173 pKa = 4.78SIDD176 pKa = 3.79EE177 pKa = 4.19DD178 pKa = 3.79WEE180 pKa = 4.55VYY182 pKa = 10.52RR183 pKa = 11.84MANSFEE189 pKa = 4.13NWVEE193 pKa = 4.31SYY195 pKa = 10.0WLGEE199 pKa = 4.17EE200 pKa = 4.32EE201 pKa = 5.24SDD203 pKa = 3.97IATLFDD209 pKa = 5.02EE210 pKa = 5.51IGQEE214 pKa = 4.06MGVLTSDD221 pKa = 4.35GEE223 pKa = 4.52FNQEE227 pKa = 3.6GYY229 pKa = 10.28FRR231 pKa = 11.84SAVHH235 pKa = 5.44EE236 pKa = 4.03HH237 pKa = 6.44LMNQFGDD244 pKa = 3.92EE245 pKa = 3.98NFMVIDD251 pKa = 3.64EE252 pKa = 4.53TEE254 pKa = 4.17YY255 pKa = 10.78PGDD258 pKa = 3.93ALYY261 pKa = 10.58DD262 pKa = 3.72IYY264 pKa = 10.84WFAPEE269 pKa = 3.91EE270 pKa = 3.88QRR272 pKa = 11.84PYY274 pKa = 10.6HH275 pKa = 6.65ALATNGLSTIEE286 pKa = 4.23MPSAEE291 pKa = 5.56DD292 pKa = 3.88GDD294 pKa = 4.12TKK296 pKa = 10.92NTFEE300 pKa = 5.47HH301 pKa = 6.79IEE303 pKa = 4.02LLAFFPPEE311 pKa = 3.72YY312 pKa = 10.09FLNGDD317 pKa = 4.0QIQMTEE323 pKa = 3.88QQSWILAVFKK333 pKa = 10.25TLAEE337 pKa = 4.04LLKK340 pKa = 9.96EE341 pKa = 4.18GQRR344 pKa = 11.84IDD346 pKa = 3.3INSTFEE352 pKa = 3.99LSGVEE357 pKa = 3.78ALPFKK362 pKa = 10.27EE363 pKa = 3.44ISLRR367 pKa = 11.84YY368 pKa = 8.25STLFTAQEE376 pKa = 4.21TLISTAHH383 pKa = 6.37GEE385 pKa = 4.4SIRR388 pKa = 11.84QLVIVPLYY396 pKa = 10.52SAEE399 pKa = 3.78QQYY402 pKa = 10.48QYY404 pKa = 10.96EE405 pKa = 4.26YY406 pKa = 11.13GYY408 pKa = 11.35AALNQTLKK416 pKa = 10.61AANVGDD422 pKa = 4.31IINPQRR428 pKa = 11.84ANAVSQQQ435 pKa = 3.05
Molecular weight: 49.86 kDa
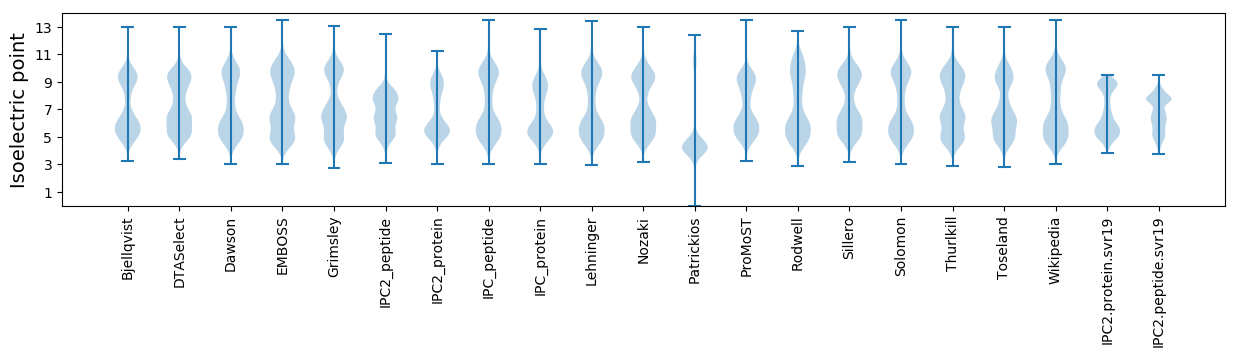
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A380RAS1|A0A380RAS1_9GAMM GTP-binding protein TypA/BipA homolog OS=Suttonella ornithocola OX=279832 GN=typA_2 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLKK12 pKa = 10.2RR13 pKa = 11.84KK14 pKa = 7.57RR15 pKa = 11.84THH17 pKa = 5.9GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.4NGRR29 pKa = 11.84QVLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.03GRR40 pKa = 11.84HH41 pKa = 5.34RR42 pKa = 11.84LTVV45 pKa = 3.07
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLKK12 pKa = 10.2RR13 pKa = 11.84KK14 pKa = 7.57RR15 pKa = 11.84THH17 pKa = 5.9GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.4NGRR29 pKa = 11.84QVLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.03GRR40 pKa = 11.84HH41 pKa = 5.34RR42 pKa = 11.84LTVV45 pKa = 3.07
Molecular weight: 5.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
711237 |
30 |
4094 |
286.4 |
31.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.147 ± 0.061 | 0.951 ± 0.017 |
5.098 ± 0.052 | 6.083 ± 0.055 |
4.005 ± 0.045 | 6.564 ± 0.057 |
2.38 ± 0.024 | 7.379 ± 0.051 |
5.62 ± 0.046 | 10.446 ± 0.071 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.357 ± 0.029 | 4.469 ± 0.046 |
4.081 ± 0.034 | 4.798 ± 0.044 |
4.76 ± 0.047 | 5.931 ± 0.048 |
5.461 ± 0.057 | 5.8 ± 0.056 |
1.315 ± 0.027 | 3.355 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
