
Apis mellifera associated microvirus 17
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 7.78
Get precalculated fractions of proteins
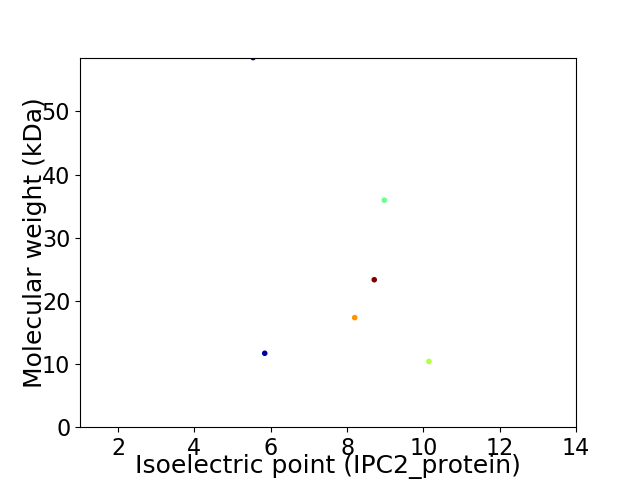
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3S8UU97|A0A3S8UU97_9VIRU DNA pilot protein OS=Apis mellifera associated microvirus 17 OX=2494744 PE=4 SV=1
MM1 pKa = 7.39KK2 pKa = 10.44SVMLHH7 pKa = 6.56DD8 pKa = 4.8FSKK11 pKa = 10.56VPHH14 pKa = 6.31VEE16 pKa = 3.72IPRR19 pKa = 11.84SSFNRR24 pKa = 11.84SHH26 pKa = 6.45GCKK29 pKa = 7.67TTFNAGYY36 pKa = 9.15LVPVFVDD43 pKa = 3.93EE44 pKa = 4.54VLPGDD49 pKa = 3.98TFNLRR54 pKa = 11.84MNVFARR60 pKa = 11.84LATPLTPFMDD70 pKa = 3.48NMFLEE75 pKa = 4.65SFFFFVPYY83 pKa = 10.21RR84 pKa = 11.84LVWNNFQKK92 pKa = 10.9FMGEE96 pKa = 3.94QEE98 pKa = 4.65DD99 pKa = 4.39PGDD102 pKa = 3.8STDD105 pKa = 3.61YY106 pKa = 11.2LIPQVSAAVGGYY118 pKa = 9.18AQNSLFDD125 pKa = 3.78YY126 pKa = 10.76FGIPTQISGANPMLPNALPFRR147 pKa = 11.84AYY149 pKa = 11.07NLIWNEE155 pKa = 3.57WFRR158 pKa = 11.84DD159 pKa = 3.65QNLQNSATVNKK170 pKa = 10.26GDD172 pKa = 4.43GPDD175 pKa = 3.58SPVATTLLRR184 pKa = 11.84RR185 pKa = 11.84GKK187 pKa = 9.54RR188 pKa = 11.84HH189 pKa = 7.42DD190 pKa = 4.07YY191 pKa = 8.21FTSCLPWPQKK201 pKa = 10.67GDD203 pKa = 3.26AVTIPLVPEE212 pKa = 4.46SSTHH216 pKa = 5.05TIYY219 pKa = 10.69RR220 pKa = 11.84RR221 pKa = 11.84ANANVWQMYY230 pKa = 8.82VAGSEE235 pKa = 4.46TIAGTTGNLTNTATNGFLNSTGAGHH260 pKa = 7.13VSMNPGDD267 pKa = 4.2GLRR270 pKa = 11.84VDD272 pKa = 4.07VLPDD276 pKa = 3.36VTGGTINEE284 pKa = 4.06LRR286 pKa = 11.84QAFQIQKK293 pKa = 10.23LYY295 pKa = 10.9EE296 pKa = 4.3RR297 pKa = 11.84DD298 pKa = 3.34ARR300 pKa = 11.84GGTRR304 pKa = 11.84YY305 pKa = 8.58TEE307 pKa = 4.72IIRR310 pKa = 11.84SHH312 pKa = 6.66FGVVSPDD319 pKa = 3.07ARR321 pKa = 11.84LQRR324 pKa = 11.84PEE326 pKa = 3.87YY327 pKa = 10.14LGGGSTPVNINPVPQTSEE345 pKa = 4.16AGTTPQGNLAATGVAQGGGHH365 pKa = 6.62GFSKK369 pKa = 10.9SFTEE373 pKa = 3.84HH374 pKa = 6.6GIVIGMVSVRR384 pKa = 11.84ADD386 pKa = 3.34LNYY389 pKa = 10.12QQGLNRR395 pKa = 11.84MWSRR399 pKa = 11.84RR400 pKa = 11.84TKK402 pKa = 10.89LDD404 pKa = 3.97FYY406 pKa = 10.76WPTLSHH412 pKa = 7.04IGEE415 pKa = 3.93QAVLNKK421 pKa = 10.07EE422 pKa = 3.95IYY424 pKa = 9.63AQEE427 pKa = 3.82TAADD431 pKa = 4.11DD432 pKa = 4.08EE433 pKa = 4.89VFGYY437 pKa = 7.47QEE439 pKa = 4.05RR440 pKa = 11.84YY441 pKa = 9.49GEE443 pKa = 4.08YY444 pKa = 9.52RR445 pKa = 11.84YY446 pKa = 10.31KK447 pKa = 10.73PSIITGDD454 pKa = 3.51FRR456 pKa = 11.84SNASTSLDD464 pKa = 3.43TWHH467 pKa = 7.46LAQDD471 pKa = 4.44FASLPTLGPTFIEE484 pKa = 4.29EE485 pKa = 4.32NPPVARR491 pKa = 11.84VIATPTEE498 pKa = 3.74PHH500 pKa = 6.35FLFDD504 pKa = 3.65SYY506 pKa = 11.84MNLTCARR513 pKa = 11.84PMPVYY518 pKa = 10.19GVPGLVDD525 pKa = 3.28HH526 pKa = 7.13FF527 pKa = 4.86
MM1 pKa = 7.39KK2 pKa = 10.44SVMLHH7 pKa = 6.56DD8 pKa = 4.8FSKK11 pKa = 10.56VPHH14 pKa = 6.31VEE16 pKa = 3.72IPRR19 pKa = 11.84SSFNRR24 pKa = 11.84SHH26 pKa = 6.45GCKK29 pKa = 7.67TTFNAGYY36 pKa = 9.15LVPVFVDD43 pKa = 3.93EE44 pKa = 4.54VLPGDD49 pKa = 3.98TFNLRR54 pKa = 11.84MNVFARR60 pKa = 11.84LATPLTPFMDD70 pKa = 3.48NMFLEE75 pKa = 4.65SFFFFVPYY83 pKa = 10.21RR84 pKa = 11.84LVWNNFQKK92 pKa = 10.9FMGEE96 pKa = 3.94QEE98 pKa = 4.65DD99 pKa = 4.39PGDD102 pKa = 3.8STDD105 pKa = 3.61YY106 pKa = 11.2LIPQVSAAVGGYY118 pKa = 9.18AQNSLFDD125 pKa = 3.78YY126 pKa = 10.76FGIPTQISGANPMLPNALPFRR147 pKa = 11.84AYY149 pKa = 11.07NLIWNEE155 pKa = 3.57WFRR158 pKa = 11.84DD159 pKa = 3.65QNLQNSATVNKK170 pKa = 10.26GDD172 pKa = 4.43GPDD175 pKa = 3.58SPVATTLLRR184 pKa = 11.84RR185 pKa = 11.84GKK187 pKa = 9.54RR188 pKa = 11.84HH189 pKa = 7.42DD190 pKa = 4.07YY191 pKa = 8.21FTSCLPWPQKK201 pKa = 10.67GDD203 pKa = 3.26AVTIPLVPEE212 pKa = 4.46SSTHH216 pKa = 5.05TIYY219 pKa = 10.69RR220 pKa = 11.84RR221 pKa = 11.84ANANVWQMYY230 pKa = 8.82VAGSEE235 pKa = 4.46TIAGTTGNLTNTATNGFLNSTGAGHH260 pKa = 7.13VSMNPGDD267 pKa = 4.2GLRR270 pKa = 11.84VDD272 pKa = 4.07VLPDD276 pKa = 3.36VTGGTINEE284 pKa = 4.06LRR286 pKa = 11.84QAFQIQKK293 pKa = 10.23LYY295 pKa = 10.9EE296 pKa = 4.3RR297 pKa = 11.84DD298 pKa = 3.34ARR300 pKa = 11.84GGTRR304 pKa = 11.84YY305 pKa = 8.58TEE307 pKa = 4.72IIRR310 pKa = 11.84SHH312 pKa = 6.66FGVVSPDD319 pKa = 3.07ARR321 pKa = 11.84LQRR324 pKa = 11.84PEE326 pKa = 3.87YY327 pKa = 10.14LGGGSTPVNINPVPQTSEE345 pKa = 4.16AGTTPQGNLAATGVAQGGGHH365 pKa = 6.62GFSKK369 pKa = 10.9SFTEE373 pKa = 3.84HH374 pKa = 6.6GIVIGMVSVRR384 pKa = 11.84ADD386 pKa = 3.34LNYY389 pKa = 10.12QQGLNRR395 pKa = 11.84MWSRR399 pKa = 11.84RR400 pKa = 11.84TKK402 pKa = 10.89LDD404 pKa = 3.97FYY406 pKa = 10.76WPTLSHH412 pKa = 7.04IGEE415 pKa = 3.93QAVLNKK421 pKa = 10.07EE422 pKa = 3.95IYY424 pKa = 9.63AQEE427 pKa = 3.82TAADD431 pKa = 4.11DD432 pKa = 4.08EE433 pKa = 4.89VFGYY437 pKa = 7.47QEE439 pKa = 4.05RR440 pKa = 11.84YY441 pKa = 9.49GEE443 pKa = 4.08YY444 pKa = 9.52RR445 pKa = 11.84YY446 pKa = 10.31KK447 pKa = 10.73PSIITGDD454 pKa = 3.51FRR456 pKa = 11.84SNASTSLDD464 pKa = 3.43TWHH467 pKa = 7.46LAQDD471 pKa = 4.44FASLPTLGPTFIEE484 pKa = 4.29EE485 pKa = 4.32NPPVARR491 pKa = 11.84VIATPTEE498 pKa = 3.74PHH500 pKa = 6.35FLFDD504 pKa = 3.65SYY506 pKa = 11.84MNLTCARR513 pKa = 11.84PMPVYY518 pKa = 10.19GVPGLVDD525 pKa = 3.28HH526 pKa = 7.13FF527 pKa = 4.86
Molecular weight: 58.51 kDa
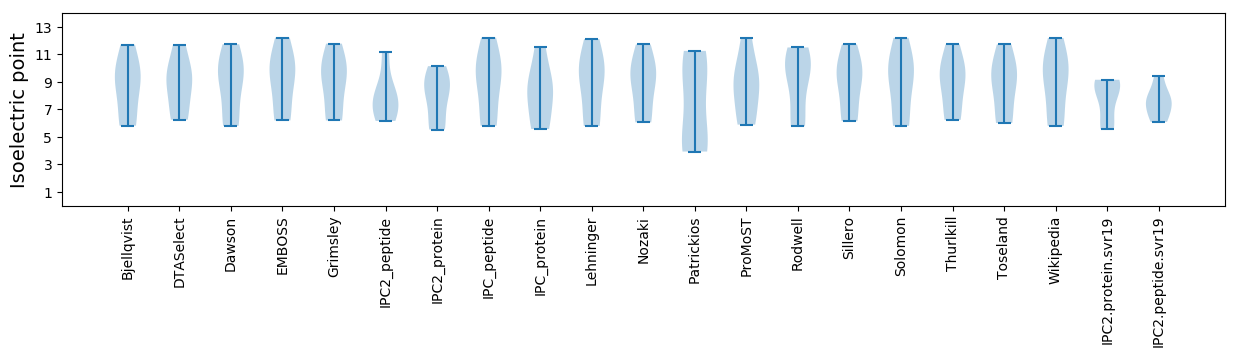
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3S8UU76|A0A3S8UU76_9VIRU Major capsid protein OS=Apis mellifera associated microvirus 17 OX=2494744 PE=3 SV=1
MM1 pKa = 7.86AKK3 pKa = 9.99PLLAQRR9 pKa = 11.84VIMRR13 pKa = 11.84DD14 pKa = 3.5DD15 pKa = 3.18LTLRR19 pKa = 11.84SLRR22 pKa = 11.84DD23 pKa = 3.21FHH25 pKa = 6.48TVLLRR30 pKa = 11.84TLGLHH35 pKa = 6.37KK36 pKa = 10.75AGALTSEE43 pKa = 4.42GWPFKK48 pKa = 10.77RR49 pKa = 11.84KK50 pKa = 6.66ITRR53 pKa = 11.84RR54 pKa = 11.84SSLQIARR61 pKa = 11.84PYY63 pKa = 10.61VLIGAQCVRR72 pKa = 11.84ATRR75 pKa = 11.84ALMSLNPAADD85 pKa = 3.8RR86 pKa = 11.84PVVPLPTT93 pKa = 3.79
MM1 pKa = 7.86AKK3 pKa = 9.99PLLAQRR9 pKa = 11.84VIMRR13 pKa = 11.84DD14 pKa = 3.5DD15 pKa = 3.18LTLRR19 pKa = 11.84SLRR22 pKa = 11.84DD23 pKa = 3.21FHH25 pKa = 6.48TVLLRR30 pKa = 11.84TLGLHH35 pKa = 6.37KK36 pKa = 10.75AGALTSEE43 pKa = 4.42GWPFKK48 pKa = 10.77RR49 pKa = 11.84KK50 pKa = 6.66ITRR53 pKa = 11.84RR54 pKa = 11.84SSLQIARR61 pKa = 11.84PYY63 pKa = 10.61VLIGAQCVRR72 pKa = 11.84ATRR75 pKa = 11.84ALMSLNPAADD85 pKa = 3.8RR86 pKa = 11.84PVVPLPTT93 pKa = 3.79
Molecular weight: 10.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1413 |
93 |
527 |
235.5 |
26.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.917 ± 1.507 | 1.132 ± 0.519 |
4.954 ± 0.306 | 5.662 ± 0.778 |
4.529 ± 0.735 | 7.289 ± 0.594 |
2.335 ± 0.405 | 4.317 ± 0.207 |
5.732 ± 1.468 | 7.643 ± 1.003 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.335 ± 0.479 | 4.6 ± 0.864 |
6.723 ± 0.753 | 4.105 ± 0.736 |
6.865 ± 1.043 | 6.44 ± 0.41 |
6.582 ± 0.677 | 5.591 ± 0.809 |
0.991 ± 0.32 | 3.255 ± 0.701 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
