
Flavivirga rizhaonensis
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Flavivirga
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
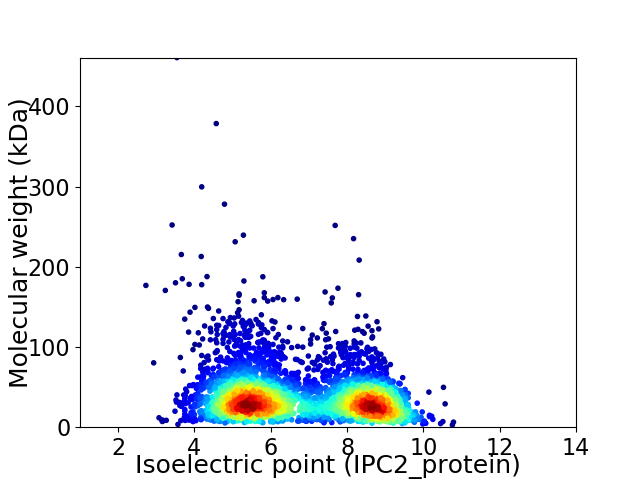
Virtual 2D-PAGE plot for 4045 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4S1E2P9|A0A4S1E2P9_9FLAO AhpC/TSA family protein OS=Flavivirga rizhaonensis OX=2559571 GN=EM932_01795 PE=4 SV=1
MM1 pKa = 7.24NKK3 pKa = 9.42IMMHH7 pKa = 6.86LLQKK11 pKa = 9.94KK12 pKa = 9.1LQMLTIVFLSSMGAMAQAEE31 pKa = 4.92YY32 pKa = 9.02ITTWNTTNAGVSNSNSIIIPAIGGRR57 pKa = 11.84YY58 pKa = 9.69DD59 pKa = 3.41VDD61 pKa = 3.14IGNDD65 pKa = 3.41GSYY68 pKa = 11.05EE69 pKa = 3.98LLNQTGRR76 pKa = 11.84ITIDD80 pKa = 3.3VTAHH84 pKa = 6.39GYY86 pKa = 8.9AAGVIQIALRR96 pKa = 11.84NAISGAGNLEE106 pKa = 3.98QIQFEE111 pKa = 4.18PSGTNTDD118 pKa = 2.89NGKK121 pKa = 9.47IVSIDD126 pKa = 2.98QWGNSIVWLSMHH138 pKa = 6.34SAYY141 pKa = 10.33HH142 pKa = 5.28SCINLDD148 pKa = 3.38VVAFDD153 pKa = 5.16APNLSQVFNMILMFANCTSMVGTPAFNTWDD183 pKa = 3.56VSNVTTTGGMFGDD196 pKa = 4.15TPFNQPLNNWDD207 pKa = 3.45VSNVTNMGFMFSNCPFNQPIGNWVTSSVTNLTGMFFNNTVFNQPIGNWDD256 pKa = 3.3VSLVTNMQGVFNGATSFNQPLNNWNVSSVIVMDD289 pKa = 4.25DD290 pKa = 3.06MFRR293 pKa = 11.84RR294 pKa = 11.84ATSFNQPLNNWDD306 pKa = 3.33VSNVVDD312 pKa = 4.06MNLMFLSSAFNQPIGNWDD330 pKa = 3.44VSSVTNMGGMFFNNTVFNQPIGSWDD355 pKa = 3.46VSSVIDD361 pKa = 3.82MNSMFNNATTFNQSLGAWNIGNLTSGVGFLDD392 pKa = 3.66NSGLSLANWDD402 pKa = 3.63ATLTGWDD409 pKa = 3.38AQGFGNPNVTIGANGLQYY427 pKa = 10.86CSAQTQRR434 pKa = 11.84DD435 pKa = 3.5AMQTAGIFTFVGDD448 pKa = 4.12VLGCTPIITCGDD460 pKa = 3.59AFNASSISKK469 pKa = 9.44FSNRR473 pKa = 11.84FEE475 pKa = 4.39VTLHH479 pKa = 6.63LDD481 pKa = 3.47APTGLAFNTDD491 pKa = 3.38GTKK494 pKa = 9.49MFAIAEE500 pKa = 4.27GLGGNDD506 pKa = 5.5DD507 pKa = 5.37IIAEE511 pKa = 4.31YY512 pKa = 10.39NLSIGFDD519 pKa = 3.42TTTAVYY525 pKa = 10.43AGAAEE530 pKa = 4.0QFFVGGQEE538 pKa = 4.1SDD540 pKa = 3.29PHH542 pKa = 7.48GLTFNAEE549 pKa = 3.97GTKK552 pKa = 9.76MFVIGLNGDD561 pKa = 3.57AVVEE565 pKa = 4.3YY566 pKa = 10.54NLGVAFDD573 pKa = 3.86VSTAIHH579 pKa = 6.55AGAAEE584 pKa = 4.1EE585 pKa = 4.36FFVGGQEE592 pKa = 4.0TSPTGLAFNTDD603 pKa = 3.42GTKK606 pKa = 9.48MFAVGVDD613 pKa = 3.47DD614 pKa = 4.52GAVVEE619 pKa = 4.79YY620 pKa = 10.73NLSTGFDD627 pKa = 3.34VSTAVYY633 pKa = 10.09AGASEE638 pKa = 4.21EE639 pKa = 4.24FFVGGQEE646 pKa = 4.2SSPTGLTFNTDD657 pKa = 2.93GTKK660 pKa = 9.66MFVIGLNGDD669 pKa = 3.57AVVEE673 pKa = 4.35YY674 pKa = 10.32NLSAGFDD681 pKa = 3.52VSTAVYY687 pKa = 10.16AGTSEE692 pKa = 4.1EE693 pKa = 4.32FSLLGYY699 pKa = 9.54SASPVDD705 pKa = 5.64LVFNTNGTKK714 pKa = 10.36LYY716 pKa = 10.97VLDD719 pKa = 4.36GLFGGNYY726 pKa = 9.35VLEE729 pKa = 4.62FVQDD733 pKa = 3.04VDD735 pKa = 3.99NYY737 pKa = 11.24HH738 pKa = 6.86EE739 pKa = 4.34ATATNGNIMANTKK752 pKa = 9.64PLVYY756 pKa = 10.13TIINDD761 pKa = 3.91TFSDD765 pKa = 3.9TNNDD769 pKa = 3.52DD770 pKa = 3.78VLDD773 pKa = 4.25GGFTISNLPSGLTAEE788 pKa = 5.07FVLSHH793 pKa = 6.83GDD795 pKa = 3.23TRR797 pKa = 11.84ATLVLSGNAANNNSSNSIATLNIVFNATAVTSNTAPTLNCSSTTGIDD844 pKa = 4.17FIDD847 pKa = 3.93SAIVCANIFDD857 pKa = 4.05VSTSEE862 pKa = 4.26FSNQFSVNAQEE873 pKa = 3.93NQLRR877 pKa = 11.84GVEE880 pKa = 4.3FNTDD884 pKa = 2.99GTKK887 pKa = 9.63MFVIGDD893 pKa = 3.63SDD895 pKa = 4.06DD896 pKa = 5.92AIVEE900 pKa = 4.47YY901 pKa = 10.86NLSTGFDD908 pKa = 3.34VSTAVYY914 pKa = 10.18AGASQEE920 pKa = 4.19FFFGGQVPNPLNFAFSTDD938 pKa = 3.21GTKK941 pKa = 10.84LFIINGIGPGGSSGLYY957 pKa = 9.63QYY959 pKa = 11.88NLGTGFDD966 pKa = 3.36VSTAVFAGGFHH977 pKa = 7.0RR978 pKa = 11.84RR979 pKa = 11.84FISQEE984 pKa = 3.9SFPTDD989 pKa = 3.09LAFNTDD995 pKa = 3.18GSKK998 pKa = 9.73MFIIGVSDD1006 pKa = 3.84DD1007 pKa = 3.95AVIEE1011 pKa = 4.33YY1012 pKa = 10.28ILRR1015 pKa = 11.84ANFDD1019 pKa = 3.4ISTMVYY1025 pKa = 10.12AGAAEE1030 pKa = 4.26EE1031 pKa = 4.26FSVVAQEE1038 pKa = 4.7SEE1040 pKa = 4.2PTGLAFNTDD1049 pKa = 3.22GTKK1052 pKa = 9.6MFVIGNSGDD1061 pKa = 3.52AVVEE1065 pKa = 4.38YY1066 pKa = 10.74NLSTGFDD1073 pKa = 3.31VSTAVYY1079 pKa = 10.01SGASEE1084 pKa = 4.26EE1085 pKa = 4.41FSVTTFEE1092 pKa = 5.14AIPRR1096 pKa = 11.84GLAFNANGSKK1106 pKa = 10.49LYY1108 pKa = 10.91VSGLVRR1114 pKa = 11.84DD1115 pKa = 4.53SIIEE1119 pKa = 4.09FAVGEE1124 pKa = 3.98ADD1126 pKa = 3.31NYY1128 pKa = 11.02EE1129 pKa = 4.03EE1130 pKa = 5.09ASINNGSIATNTSPFIYY1147 pKa = 10.1NVSNDD1152 pKa = 2.97TFADD1156 pKa = 3.73VNNDD1160 pKa = 4.03DD1161 pKa = 4.38ILDD1164 pKa = 3.94GGFTIGNLSPGLTPQFVLSNGDD1186 pKa = 3.25TTATLVLSGNATNNSSSDD1204 pKa = 3.59NIADD1208 pKa = 4.06LNISFNTTAITSTTVPILDD1227 pKa = 4.36CNTIVGIDD1235 pKa = 3.67FLDD1238 pKa = 4.27NINLNGRR1245 pKa = 11.84LRR1247 pKa = 11.84HH1248 pKa = 6.02GKK1250 pKa = 9.64FFLNGSEE1257 pKa = 4.03QPMVTGRR1264 pKa = 11.84NANN1267 pKa = 3.36
MM1 pKa = 7.24NKK3 pKa = 9.42IMMHH7 pKa = 6.86LLQKK11 pKa = 9.94KK12 pKa = 9.1LQMLTIVFLSSMGAMAQAEE31 pKa = 4.92YY32 pKa = 9.02ITTWNTTNAGVSNSNSIIIPAIGGRR57 pKa = 11.84YY58 pKa = 9.69DD59 pKa = 3.41VDD61 pKa = 3.14IGNDD65 pKa = 3.41GSYY68 pKa = 11.05EE69 pKa = 3.98LLNQTGRR76 pKa = 11.84ITIDD80 pKa = 3.3VTAHH84 pKa = 6.39GYY86 pKa = 8.9AAGVIQIALRR96 pKa = 11.84NAISGAGNLEE106 pKa = 3.98QIQFEE111 pKa = 4.18PSGTNTDD118 pKa = 2.89NGKK121 pKa = 9.47IVSIDD126 pKa = 2.98QWGNSIVWLSMHH138 pKa = 6.34SAYY141 pKa = 10.33HH142 pKa = 5.28SCINLDD148 pKa = 3.38VVAFDD153 pKa = 5.16APNLSQVFNMILMFANCTSMVGTPAFNTWDD183 pKa = 3.56VSNVTTTGGMFGDD196 pKa = 4.15TPFNQPLNNWDD207 pKa = 3.45VSNVTNMGFMFSNCPFNQPIGNWVTSSVTNLTGMFFNNTVFNQPIGNWDD256 pKa = 3.3VSLVTNMQGVFNGATSFNQPLNNWNVSSVIVMDD289 pKa = 4.25DD290 pKa = 3.06MFRR293 pKa = 11.84RR294 pKa = 11.84ATSFNQPLNNWDD306 pKa = 3.33VSNVVDD312 pKa = 4.06MNLMFLSSAFNQPIGNWDD330 pKa = 3.44VSSVTNMGGMFFNNTVFNQPIGSWDD355 pKa = 3.46VSSVIDD361 pKa = 3.82MNSMFNNATTFNQSLGAWNIGNLTSGVGFLDD392 pKa = 3.66NSGLSLANWDD402 pKa = 3.63ATLTGWDD409 pKa = 3.38AQGFGNPNVTIGANGLQYY427 pKa = 10.86CSAQTQRR434 pKa = 11.84DD435 pKa = 3.5AMQTAGIFTFVGDD448 pKa = 4.12VLGCTPIITCGDD460 pKa = 3.59AFNASSISKK469 pKa = 9.44FSNRR473 pKa = 11.84FEE475 pKa = 4.39VTLHH479 pKa = 6.63LDD481 pKa = 3.47APTGLAFNTDD491 pKa = 3.38GTKK494 pKa = 9.49MFAIAEE500 pKa = 4.27GLGGNDD506 pKa = 5.5DD507 pKa = 5.37IIAEE511 pKa = 4.31YY512 pKa = 10.39NLSIGFDD519 pKa = 3.42TTTAVYY525 pKa = 10.43AGAAEE530 pKa = 4.0QFFVGGQEE538 pKa = 4.1SDD540 pKa = 3.29PHH542 pKa = 7.48GLTFNAEE549 pKa = 3.97GTKK552 pKa = 9.76MFVIGLNGDD561 pKa = 3.57AVVEE565 pKa = 4.3YY566 pKa = 10.54NLGVAFDD573 pKa = 3.86VSTAIHH579 pKa = 6.55AGAAEE584 pKa = 4.1EE585 pKa = 4.36FFVGGQEE592 pKa = 4.0TSPTGLAFNTDD603 pKa = 3.42GTKK606 pKa = 9.48MFAVGVDD613 pKa = 3.47DD614 pKa = 4.52GAVVEE619 pKa = 4.79YY620 pKa = 10.73NLSTGFDD627 pKa = 3.34VSTAVYY633 pKa = 10.09AGASEE638 pKa = 4.21EE639 pKa = 4.24FFVGGQEE646 pKa = 4.2SSPTGLTFNTDD657 pKa = 2.93GTKK660 pKa = 9.66MFVIGLNGDD669 pKa = 3.57AVVEE673 pKa = 4.35YY674 pKa = 10.32NLSAGFDD681 pKa = 3.52VSTAVYY687 pKa = 10.16AGTSEE692 pKa = 4.1EE693 pKa = 4.32FSLLGYY699 pKa = 9.54SASPVDD705 pKa = 5.64LVFNTNGTKK714 pKa = 10.36LYY716 pKa = 10.97VLDD719 pKa = 4.36GLFGGNYY726 pKa = 9.35VLEE729 pKa = 4.62FVQDD733 pKa = 3.04VDD735 pKa = 3.99NYY737 pKa = 11.24HH738 pKa = 6.86EE739 pKa = 4.34ATATNGNIMANTKK752 pKa = 9.64PLVYY756 pKa = 10.13TIINDD761 pKa = 3.91TFSDD765 pKa = 3.9TNNDD769 pKa = 3.52DD770 pKa = 3.78VLDD773 pKa = 4.25GGFTISNLPSGLTAEE788 pKa = 5.07FVLSHH793 pKa = 6.83GDD795 pKa = 3.23TRR797 pKa = 11.84ATLVLSGNAANNNSSNSIATLNIVFNATAVTSNTAPTLNCSSTTGIDD844 pKa = 4.17FIDD847 pKa = 3.93SAIVCANIFDD857 pKa = 4.05VSTSEE862 pKa = 4.26FSNQFSVNAQEE873 pKa = 3.93NQLRR877 pKa = 11.84GVEE880 pKa = 4.3FNTDD884 pKa = 2.99GTKK887 pKa = 9.63MFVIGDD893 pKa = 3.63SDD895 pKa = 4.06DD896 pKa = 5.92AIVEE900 pKa = 4.47YY901 pKa = 10.86NLSTGFDD908 pKa = 3.34VSTAVYY914 pKa = 10.18AGASQEE920 pKa = 4.19FFFGGQVPNPLNFAFSTDD938 pKa = 3.21GTKK941 pKa = 10.84LFIINGIGPGGSSGLYY957 pKa = 9.63QYY959 pKa = 11.88NLGTGFDD966 pKa = 3.36VSTAVFAGGFHH977 pKa = 7.0RR978 pKa = 11.84RR979 pKa = 11.84FISQEE984 pKa = 3.9SFPTDD989 pKa = 3.09LAFNTDD995 pKa = 3.18GSKK998 pKa = 9.73MFIIGVSDD1006 pKa = 3.84DD1007 pKa = 3.95AVIEE1011 pKa = 4.33YY1012 pKa = 10.28ILRR1015 pKa = 11.84ANFDD1019 pKa = 3.4ISTMVYY1025 pKa = 10.12AGAAEE1030 pKa = 4.26EE1031 pKa = 4.26FSVVAQEE1038 pKa = 4.7SEE1040 pKa = 4.2PTGLAFNTDD1049 pKa = 3.22GTKK1052 pKa = 9.6MFVIGNSGDD1061 pKa = 3.52AVVEE1065 pKa = 4.38YY1066 pKa = 10.74NLSTGFDD1073 pKa = 3.31VSTAVYY1079 pKa = 10.01SGASEE1084 pKa = 4.26EE1085 pKa = 4.41FSVTTFEE1092 pKa = 5.14AIPRR1096 pKa = 11.84GLAFNANGSKK1106 pKa = 10.49LYY1108 pKa = 10.91VSGLVRR1114 pKa = 11.84DD1115 pKa = 4.53SIIEE1119 pKa = 4.09FAVGEE1124 pKa = 3.98ADD1126 pKa = 3.31NYY1128 pKa = 11.02EE1129 pKa = 4.03EE1130 pKa = 5.09ASINNGSIATNTSPFIYY1147 pKa = 10.1NVSNDD1152 pKa = 2.97TFADD1156 pKa = 3.73VNNDD1160 pKa = 4.03DD1161 pKa = 4.38ILDD1164 pKa = 3.94GGFTIGNLSPGLTPQFVLSNGDD1186 pKa = 3.25TTATLVLSGNATNNSSSDD1204 pKa = 3.59NIADD1208 pKa = 4.06LNISFNTTAITSTTVPILDD1227 pKa = 4.36CNTIVGIDD1235 pKa = 3.67FLDD1238 pKa = 4.27NINLNGRR1245 pKa = 11.84LRR1247 pKa = 11.84HH1248 pKa = 6.02GKK1250 pKa = 9.64FFLNGSEE1257 pKa = 4.03QPMVTGRR1264 pKa = 11.84NANN1267 pKa = 3.36
Molecular weight: 134.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4S1DXL5|A0A4S1DXL5_9FLAO SRPBCC domain-containing protein OS=Flavivirga rizhaonensis OX=2559571 GN=EM932_08855 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.76GRR40 pKa = 11.84KK41 pKa = 7.91RR42 pKa = 11.84LSVSSEE48 pKa = 3.87TRR50 pKa = 11.84HH51 pKa = 5.96KK52 pKa = 10.68KK53 pKa = 9.8
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSKK10 pKa = 9.13RR11 pKa = 11.84KK12 pKa = 9.48RR13 pKa = 11.84RR14 pKa = 11.84NKK16 pKa = 9.49HH17 pKa = 3.94GFRR20 pKa = 11.84EE21 pKa = 4.27RR22 pKa = 11.84MASANGRR29 pKa = 11.84KK30 pKa = 9.04VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 9.76GRR40 pKa = 11.84KK41 pKa = 7.91RR42 pKa = 11.84LSVSSEE48 pKa = 3.87TRR50 pKa = 11.84HH51 pKa = 5.96KK52 pKa = 10.68KK53 pKa = 9.8
Molecular weight: 6.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1423285 |
23 |
4414 |
351.9 |
39.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.829 ± 0.035 | 0.778 ± 0.013 |
5.732 ± 0.03 | 6.31 ± 0.039 |
5.256 ± 0.029 | 6.266 ± 0.039 |
1.8 ± 0.016 | 8.377 ± 0.037 |
8.195 ± 0.056 | 9.168 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.005 ± 0.019 | 6.753 ± 0.043 |
3.289 ± 0.025 | 3.104 ± 0.021 |
3.303 ± 0.024 | 6.701 ± 0.028 |
5.881 ± 0.047 | 5.967 ± 0.025 |
1.083 ± 0.014 | 4.2 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
