
Aquimonas voraii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Rhodanobacteraceae; Aquimonas
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
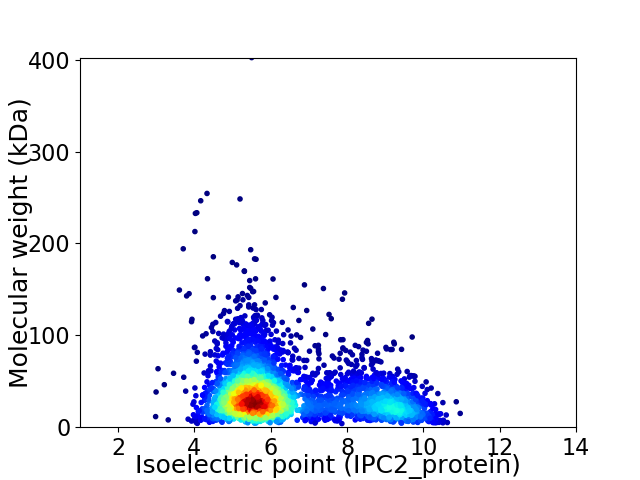
Virtual 2D-PAGE plot for 3593 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
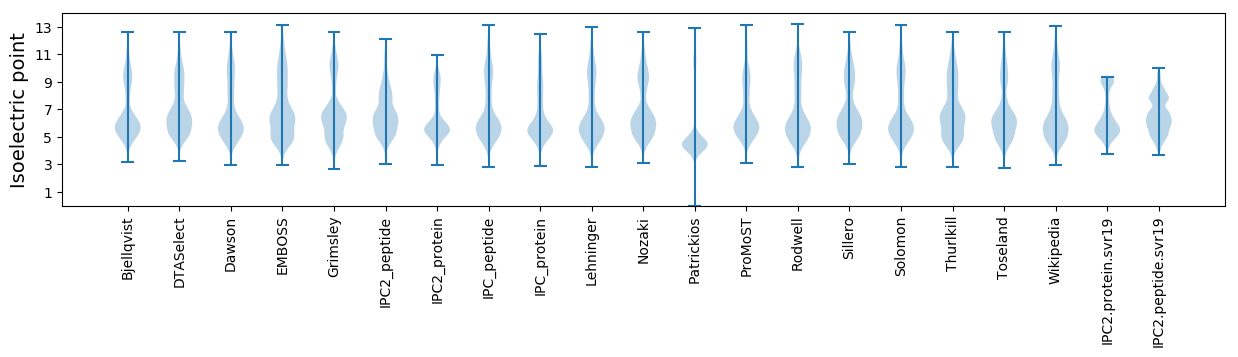
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1G6X711|A0A1G6X711_9GAMM DNA polymerase IV OS=Aquimonas voraii OX=265719 GN=dinB PE=3 SV=1
MM1 pKa = 7.68RR2 pKa = 11.84RR3 pKa = 11.84QSPRR7 pKa = 11.84KK8 pKa = 9.59SLVWLCGSLIGGLGAWSALPAQNAAALEE36 pKa = 4.19YY37 pKa = 10.14EE38 pKa = 4.21FAEE41 pKa = 4.0IHH43 pKa = 5.61EE44 pKa = 4.98AEE46 pKa = 4.46ALAPPGKK53 pKa = 9.61PGAPSMGSPLGAAPALLGMGFDD75 pKa = 5.88GEE77 pKa = 4.4IEE79 pKa = 4.41PNNSSATASPLAEE92 pKa = 3.79RR93 pKa = 11.84QGVVRR98 pKa = 11.84AKK100 pKa = 9.98LWPAADD106 pKa = 3.18IDD108 pKa = 4.18VFAIAAQAGDD118 pKa = 3.59RR119 pKa = 11.84LYY121 pKa = 11.35AGVMTSASAGSGTDD135 pKa = 2.95SLLQVIAPDD144 pKa = 3.47GTTVIEE150 pKa = 4.57ADD152 pKa = 4.01DD153 pKa = 4.17NDD155 pKa = 3.8GSFAGTSSGVAGTTLTSAGTYY176 pKa = 8.59YY177 pKa = 10.78LQVSSPSATATVRR190 pKa = 11.84PYY192 pKa = 9.98EE193 pKa = 3.76LHH195 pKa = 6.25YY196 pKa = 10.44RR197 pKa = 11.84LQAGSPTAEE206 pKa = 3.94VEE208 pKa = 4.42PNDD211 pKa = 3.74TAATANPLPASGWVSGARR229 pKa = 11.84GIAAATEE236 pKa = 3.99QDD238 pKa = 3.84WYY240 pKa = 11.44SLSLNAGDD248 pKa = 4.3TVFLSLDD255 pKa = 3.83LDD257 pKa = 3.91PEE259 pKa = 4.41RR260 pKa = 11.84DD261 pKa = 3.49NVQWDD266 pKa = 3.41GRR268 pKa = 11.84LGLGLFGDD276 pKa = 4.81ASNQILVANDD286 pKa = 3.25TSTGSVANPLSEE298 pKa = 4.16TLFITVKK305 pKa = 10.42EE306 pKa = 4.01AGTYY310 pKa = 9.58FAFVDD315 pKa = 3.87SANAAVGGPTATYY328 pKa = 9.03QLSVSVQPAANEE340 pKa = 4.06GLNCTTYY347 pKa = 10.54TSTDD351 pKa = 3.28VPVAIGPNPGLATSTITIPGNPRR374 pKa = 11.84IADD377 pKa = 3.85LDD379 pKa = 3.66VSLQLNHH386 pKa = 6.79AFMNDD391 pKa = 1.87IDD393 pKa = 4.19AQLVSPAGNSVGLFTDD409 pKa = 3.63IGATSAGGQAQMDD422 pKa = 4.0TVFDD426 pKa = 4.41DD427 pKa = 4.27EE428 pKa = 4.82AAIPPSFTALRR439 pKa = 11.84GLALKK444 pKa = 10.61PEE446 pKa = 4.43LNYY449 pKa = 10.71RR450 pKa = 11.84LAWFDD455 pKa = 3.36GTDD458 pKa = 4.27AGGTWTLEE466 pKa = 3.94LRR468 pKa = 11.84DD469 pKa = 4.61DD470 pKa = 4.72ANTDD474 pKa = 3.35GGNLTGWSLRR484 pKa = 11.84VCEE487 pKa = 4.86APPPPTCAAGFTPVTVYY504 pKa = 9.84STDD507 pKa = 3.8FEE509 pKa = 5.68AGAAGFTSSGIANEE523 pKa = 4.2WEE525 pKa = 4.18LGLPATAGTTTTNPIAAFSSCASGSSCWKK554 pKa = 8.72TDD556 pKa = 3.52LDD558 pKa = 3.8GTYY561 pKa = 10.6DD562 pKa = 3.63LSSSQDD568 pKa = 3.64LLSPNIDD575 pKa = 3.25LSGLQPPVLVRR586 pKa = 11.84WSQRR590 pKa = 11.84YY591 pKa = 7.35QMEE594 pKa = 4.17SANFDD599 pKa = 3.66RR600 pKa = 11.84YY601 pKa = 10.35HH602 pKa = 7.46VDD604 pKa = 3.66FRR606 pKa = 11.84QVGSGSGLRR615 pKa = 11.84LFEE618 pKa = 4.15WADD621 pKa = 3.07ATMANAVGNPTLQIPAAAGWSQKK644 pKa = 8.78LVRR647 pKa = 11.84ADD649 pKa = 3.66ALAGQNTEE657 pKa = 4.76LLFHH661 pKa = 7.19LDD663 pKa = 2.91SDD665 pKa = 4.17TTVNFAGVAIDD676 pKa = 4.18DD677 pKa = 4.24VSVTACRR684 pKa = 11.84ALSADD689 pKa = 3.89LAITKK694 pKa = 9.44TNGSTTSVPGGSTTYY709 pKa = 10.52TITSSNAGPDD719 pKa = 3.49AVTGATVTDD728 pKa = 4.17TFPAALTCSWTCVGAGGGTCTAASSGNLNDD758 pKa = 4.78SVNLPAGASVTHH770 pKa = 6.77TATCAIAAAATGTLSNTATISSTITDD796 pKa = 3.73PTPANNSATDD806 pKa = 3.56SDD808 pKa = 4.4TLSPQADD815 pKa = 3.63LSITKK820 pKa = 9.51TDD822 pKa = 3.24GQTAVPAGGSTVYY835 pKa = 10.62TITASNAGPSDD846 pKa = 3.86APGSTVADD854 pKa = 4.09TLPAACTSVSWTCVAAGGGTCTAAGSGNIGDD885 pKa = 4.64AANLPAGASVSYY897 pKa = 9.05TASCAIAAAASGTLSNTATVSSAVTDD923 pKa = 3.68PTPGNNAATDD933 pKa = 3.82SSSVLPPGALGASPSTLSFGTVSVGATSASQTVTLSNSGGSPLTIASLGAAAAPFALVGGSCGAVPITLAAGASCTLAYY1012 pKa = 10.48SFSPSAAGLATQSLTVNAGSAGSASLALSGIGGVGSLGLLSSSLDD1057 pKa = 3.5FGRR1060 pKa = 11.84VQVGAGGQLSLTLTNTGSGGLDD1082 pKa = 3.18VTGLSAVSAPFAQITGGSCGVVPFSLAAGTSCTVLYY1118 pKa = 10.48RR1119 pKa = 11.84FAPTATGSFNQTVTVSSTAGSATANLVGIGFVVTAVDD1156 pKa = 3.75AMDD1159 pKa = 4.05ARR1161 pKa = 11.84SLSLLALILGLIGWVTLRR1179 pKa = 11.84RR1180 pKa = 11.84RR1181 pKa = 11.84GG1182 pKa = 3.37
MM1 pKa = 7.68RR2 pKa = 11.84RR3 pKa = 11.84QSPRR7 pKa = 11.84KK8 pKa = 9.59SLVWLCGSLIGGLGAWSALPAQNAAALEE36 pKa = 4.19YY37 pKa = 10.14EE38 pKa = 4.21FAEE41 pKa = 4.0IHH43 pKa = 5.61EE44 pKa = 4.98AEE46 pKa = 4.46ALAPPGKK53 pKa = 9.61PGAPSMGSPLGAAPALLGMGFDD75 pKa = 5.88GEE77 pKa = 4.4IEE79 pKa = 4.41PNNSSATASPLAEE92 pKa = 3.79RR93 pKa = 11.84QGVVRR98 pKa = 11.84AKK100 pKa = 9.98LWPAADD106 pKa = 3.18IDD108 pKa = 4.18VFAIAAQAGDD118 pKa = 3.59RR119 pKa = 11.84LYY121 pKa = 11.35AGVMTSASAGSGTDD135 pKa = 2.95SLLQVIAPDD144 pKa = 3.47GTTVIEE150 pKa = 4.57ADD152 pKa = 4.01DD153 pKa = 4.17NDD155 pKa = 3.8GSFAGTSSGVAGTTLTSAGTYY176 pKa = 8.59YY177 pKa = 10.78LQVSSPSATATVRR190 pKa = 11.84PYY192 pKa = 9.98EE193 pKa = 3.76LHH195 pKa = 6.25YY196 pKa = 10.44RR197 pKa = 11.84LQAGSPTAEE206 pKa = 3.94VEE208 pKa = 4.42PNDD211 pKa = 3.74TAATANPLPASGWVSGARR229 pKa = 11.84GIAAATEE236 pKa = 3.99QDD238 pKa = 3.84WYY240 pKa = 11.44SLSLNAGDD248 pKa = 4.3TVFLSLDD255 pKa = 3.83LDD257 pKa = 3.91PEE259 pKa = 4.41RR260 pKa = 11.84DD261 pKa = 3.49NVQWDD266 pKa = 3.41GRR268 pKa = 11.84LGLGLFGDD276 pKa = 4.81ASNQILVANDD286 pKa = 3.25TSTGSVANPLSEE298 pKa = 4.16TLFITVKK305 pKa = 10.42EE306 pKa = 4.01AGTYY310 pKa = 9.58FAFVDD315 pKa = 3.87SANAAVGGPTATYY328 pKa = 9.03QLSVSVQPAANEE340 pKa = 4.06GLNCTTYY347 pKa = 10.54TSTDD351 pKa = 3.28VPVAIGPNPGLATSTITIPGNPRR374 pKa = 11.84IADD377 pKa = 3.85LDD379 pKa = 3.66VSLQLNHH386 pKa = 6.79AFMNDD391 pKa = 1.87IDD393 pKa = 4.19AQLVSPAGNSVGLFTDD409 pKa = 3.63IGATSAGGQAQMDD422 pKa = 4.0TVFDD426 pKa = 4.41DD427 pKa = 4.27EE428 pKa = 4.82AAIPPSFTALRR439 pKa = 11.84GLALKK444 pKa = 10.61PEE446 pKa = 4.43LNYY449 pKa = 10.71RR450 pKa = 11.84LAWFDD455 pKa = 3.36GTDD458 pKa = 4.27AGGTWTLEE466 pKa = 3.94LRR468 pKa = 11.84DD469 pKa = 4.61DD470 pKa = 4.72ANTDD474 pKa = 3.35GGNLTGWSLRR484 pKa = 11.84VCEE487 pKa = 4.86APPPPTCAAGFTPVTVYY504 pKa = 9.84STDD507 pKa = 3.8FEE509 pKa = 5.68AGAAGFTSSGIANEE523 pKa = 4.2WEE525 pKa = 4.18LGLPATAGTTTTNPIAAFSSCASGSSCWKK554 pKa = 8.72TDD556 pKa = 3.52LDD558 pKa = 3.8GTYY561 pKa = 10.6DD562 pKa = 3.63LSSSQDD568 pKa = 3.64LLSPNIDD575 pKa = 3.25LSGLQPPVLVRR586 pKa = 11.84WSQRR590 pKa = 11.84YY591 pKa = 7.35QMEE594 pKa = 4.17SANFDD599 pKa = 3.66RR600 pKa = 11.84YY601 pKa = 10.35HH602 pKa = 7.46VDD604 pKa = 3.66FRR606 pKa = 11.84QVGSGSGLRR615 pKa = 11.84LFEE618 pKa = 4.15WADD621 pKa = 3.07ATMANAVGNPTLQIPAAAGWSQKK644 pKa = 8.78LVRR647 pKa = 11.84ADD649 pKa = 3.66ALAGQNTEE657 pKa = 4.76LLFHH661 pKa = 7.19LDD663 pKa = 2.91SDD665 pKa = 4.17TTVNFAGVAIDD676 pKa = 4.18DD677 pKa = 4.24VSVTACRR684 pKa = 11.84ALSADD689 pKa = 3.89LAITKK694 pKa = 9.44TNGSTTSVPGGSTTYY709 pKa = 10.52TITSSNAGPDD719 pKa = 3.49AVTGATVTDD728 pKa = 4.17TFPAALTCSWTCVGAGGGTCTAASSGNLNDD758 pKa = 4.78SVNLPAGASVTHH770 pKa = 6.77TATCAIAAAATGTLSNTATISSTITDD796 pKa = 3.73PTPANNSATDD806 pKa = 3.56SDD808 pKa = 4.4TLSPQADD815 pKa = 3.63LSITKK820 pKa = 9.51TDD822 pKa = 3.24GQTAVPAGGSTVYY835 pKa = 10.62TITASNAGPSDD846 pKa = 3.86APGSTVADD854 pKa = 4.09TLPAACTSVSWTCVAAGGGTCTAAGSGNIGDD885 pKa = 4.64AANLPAGASVSYY897 pKa = 9.05TASCAIAAAASGTLSNTATVSSAVTDD923 pKa = 3.68PTPGNNAATDD933 pKa = 3.82SSSVLPPGALGASPSTLSFGTVSVGATSASQTVTLSNSGGSPLTIASLGAAAAPFALVGGSCGAVPITLAAGASCTLAYY1012 pKa = 10.48SFSPSAAGLATQSLTVNAGSAGSASLALSGIGGVGSLGLLSSSLDD1057 pKa = 3.5FGRR1060 pKa = 11.84VQVGAGGQLSLTLTNTGSGGLDD1082 pKa = 3.18VTGLSAVSAPFAQITGGSCGVVPFSLAAGTSCTVLYY1118 pKa = 10.48RR1119 pKa = 11.84FAPTATGSFNQTVTVSSTAGSATANLVGIGFVVTAVDD1156 pKa = 3.75AMDD1159 pKa = 4.05ARR1161 pKa = 11.84SLSLLALILGLIGWVTLRR1179 pKa = 11.84RR1180 pKa = 11.84RR1181 pKa = 11.84GG1182 pKa = 3.37
Molecular weight: 117.45 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1G6YWL0|A0A1G6YWL0_9GAMM Uncharacterized protein OS=Aquimonas voraii OX=265719 GN=SAMN04488509_11168 PE=4 SV=1
MM1 pKa = 7.69LIAKK5 pKa = 9.04PVGRR9 pKa = 11.84VFARR13 pKa = 11.84EE14 pKa = 3.4RR15 pKa = 11.84GRR17 pKa = 11.84RR18 pKa = 11.84HH19 pKa = 5.25HH20 pKa = 6.81HH21 pKa = 5.92PLRR24 pKa = 11.84LRR26 pKa = 11.84RR27 pKa = 11.84PPPRR31 pKa = 11.84HH32 pKa = 5.34RR33 pKa = 11.84HH34 pKa = 5.01RR35 pKa = 11.84PPDD38 pKa = 3.08RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84GLCLQRR48 pKa = 11.84RR49 pKa = 11.84GRR51 pKa = 11.84AARR54 pKa = 11.84AHH56 pKa = 6.39RR57 pKa = 11.84PRR59 pKa = 11.84RR60 pKa = 11.84PAHH63 pKa = 6.79RR64 pKa = 11.84DD65 pKa = 2.86RR66 pKa = 11.84ARR68 pKa = 11.84RR69 pKa = 11.84PGSHH73 pKa = 6.11HH74 pKa = 6.75RR75 pKa = 11.84APLRR79 pKa = 11.84RR80 pKa = 11.84SPKK83 pKa = 9.72RR84 pKa = 11.84RR85 pKa = 11.84GQPRR89 pKa = 11.84GLYY92 pKa = 10.17AGWRR96 pKa = 11.84RR97 pKa = 11.84PANHH101 pKa = 7.13AEE103 pKa = 3.83PVRR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84PAPRR114 pKa = 11.84AAPLRR119 pKa = 11.84RR120 pKa = 11.84PGPPDD125 pKa = 3.12RR126 pKa = 11.84RR127 pKa = 11.84RR128 pKa = 11.84RR129 pKa = 11.84PLQPAQHH136 pKa = 6.64LDD138 pKa = 3.46LRR140 pKa = 11.84RR141 pKa = 11.84SRR143 pKa = 11.84QPPEE147 pKa = 3.48RR148 pKa = 11.84DD149 pKa = 3.34GPGRR153 pKa = 11.84HH154 pKa = 5.08HH155 pKa = 6.76HH156 pKa = 6.49HH157 pKa = 7.03RR158 pKa = 11.84AGSPQPHH165 pKa = 6.83RR166 pKa = 11.84PPDD169 pKa = 3.53HH170 pKa = 6.77GRR172 pKa = 11.84GQHH175 pKa = 6.22GAQLQQRR182 pKa = 11.84RR183 pKa = 11.84PPATDD188 pKa = 3.27HH189 pKa = 6.1PPEE192 pKa = 4.51RR193 pKa = 11.84RR194 pKa = 11.84AQRR197 pKa = 11.84NPLRR201 pKa = 11.84RR202 pKa = 11.84RR203 pKa = 11.84RR204 pKa = 11.84PHH206 pKa = 6.72RR207 pKa = 11.84AHH209 pKa = 6.87HH210 pKa = 6.45PPPGQQRR217 pKa = 11.84SRR219 pKa = 11.84ALRR222 pKa = 11.84IRR224 pKa = 11.84LRR226 pKa = 11.84RR227 pKa = 11.84ARR229 pKa = 11.84QPTT232 pKa = 3.36
MM1 pKa = 7.69LIAKK5 pKa = 9.04PVGRR9 pKa = 11.84VFARR13 pKa = 11.84EE14 pKa = 3.4RR15 pKa = 11.84GRR17 pKa = 11.84RR18 pKa = 11.84HH19 pKa = 5.25HH20 pKa = 6.81HH21 pKa = 5.92PLRR24 pKa = 11.84LRR26 pKa = 11.84RR27 pKa = 11.84PPPRR31 pKa = 11.84HH32 pKa = 5.34RR33 pKa = 11.84HH34 pKa = 5.01RR35 pKa = 11.84PPDD38 pKa = 3.08RR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84GLCLQRR48 pKa = 11.84RR49 pKa = 11.84GRR51 pKa = 11.84AARR54 pKa = 11.84AHH56 pKa = 6.39RR57 pKa = 11.84PRR59 pKa = 11.84RR60 pKa = 11.84PAHH63 pKa = 6.79RR64 pKa = 11.84DD65 pKa = 2.86RR66 pKa = 11.84ARR68 pKa = 11.84RR69 pKa = 11.84PGSHH73 pKa = 6.11HH74 pKa = 6.75RR75 pKa = 11.84APLRR79 pKa = 11.84RR80 pKa = 11.84SPKK83 pKa = 9.72RR84 pKa = 11.84RR85 pKa = 11.84GQPRR89 pKa = 11.84GLYY92 pKa = 10.17AGWRR96 pKa = 11.84RR97 pKa = 11.84PANHH101 pKa = 7.13AEE103 pKa = 3.83PVRR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84PAPRR114 pKa = 11.84AAPLRR119 pKa = 11.84RR120 pKa = 11.84PGPPDD125 pKa = 3.12RR126 pKa = 11.84RR127 pKa = 11.84RR128 pKa = 11.84RR129 pKa = 11.84PLQPAQHH136 pKa = 6.64LDD138 pKa = 3.46LRR140 pKa = 11.84RR141 pKa = 11.84SRR143 pKa = 11.84QPPEE147 pKa = 3.48RR148 pKa = 11.84DD149 pKa = 3.34GPGRR153 pKa = 11.84HH154 pKa = 5.08HH155 pKa = 6.76HH156 pKa = 6.49HH157 pKa = 7.03RR158 pKa = 11.84AGSPQPHH165 pKa = 6.83RR166 pKa = 11.84PPDD169 pKa = 3.53HH170 pKa = 6.77GRR172 pKa = 11.84GQHH175 pKa = 6.22GAQLQQRR182 pKa = 11.84RR183 pKa = 11.84PPATDD188 pKa = 3.27HH189 pKa = 6.1PPEE192 pKa = 4.51RR193 pKa = 11.84RR194 pKa = 11.84AQRR197 pKa = 11.84NPLRR201 pKa = 11.84RR202 pKa = 11.84RR203 pKa = 11.84RR204 pKa = 11.84PHH206 pKa = 6.72RR207 pKa = 11.84AHH209 pKa = 6.87HH210 pKa = 6.45PPPGQQRR217 pKa = 11.84SRR219 pKa = 11.84ALRR222 pKa = 11.84IRR224 pKa = 11.84LRR226 pKa = 11.84RR227 pKa = 11.84ARR229 pKa = 11.84QPTT232 pKa = 3.36
Molecular weight: 27.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1297485 |
39 |
3779 |
361.1 |
39.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.453 ± 0.058 | 0.906 ± 0.013 |
5.336 ± 0.029 | 6.103 ± 0.044 |
3.407 ± 0.025 | 8.641 ± 0.044 |
2.028 ± 0.019 | 3.95 ± 0.029 |
2.126 ± 0.029 | 11.743 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.842 ± 0.017 | 2.318 ± 0.028 |
5.483 ± 0.031 | 3.76 ± 0.027 |
8.237 ± 0.054 | 5.699 ± 0.04 |
4.462 ± 0.049 | 7.043 ± 0.036 |
1.442 ± 0.017 | 2.022 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
