
Yellowstone Lake virophage 6
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Preplasmiviricota; Maveriviricetes; Priklausovirales; Lavidaviridae; unclassified Lavidaviridae
Average proteome isoelectric point is 7.69
Get precalculated fractions of proteins
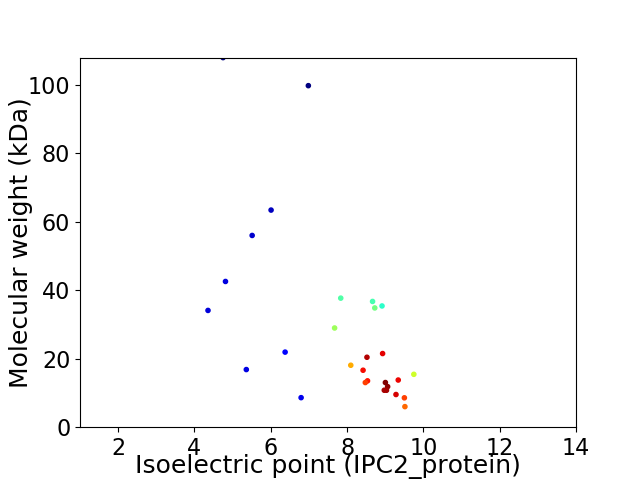
Virtual 2D-PAGE plot for 29 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A0RK71|A0A0A0RK71_9VIRU Putative primase-helicase OS=Yellowstone Lake virophage 6 OX=1557034 GN=YSLV6_ORF03 PE=4 SV=1
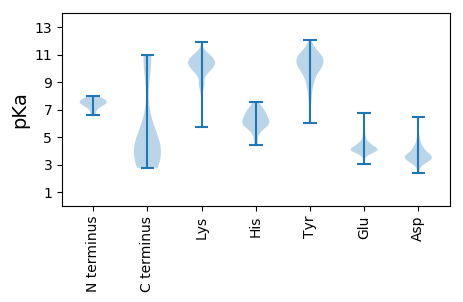
MM1 pKa = 8.02PDD3 pKa = 3.55LVTPEE8 pKa = 3.91YY9 pKa = 10.89QNYY12 pKa = 7.56TFKK15 pKa = 11.02KK16 pKa = 9.57DD17 pKa = 3.66LIGAFTLEE25 pKa = 4.15EE26 pKa = 4.12LQSEE30 pKa = 4.43FQEE33 pKa = 3.99ISNEE37 pKa = 3.68FFRR40 pKa = 11.84IKK42 pKa = 10.51EE43 pKa = 4.0LIYY46 pKa = 11.06DD47 pKa = 3.87NLEE50 pKa = 3.93EE51 pKa = 4.3SPEE54 pKa = 3.98IVVPEE59 pKa = 4.38MGLEE63 pKa = 4.25DD64 pKa = 4.72PDD66 pKa = 5.64FIDD69 pKa = 4.43KK70 pKa = 10.69LVYY73 pKa = 10.83DD74 pKa = 4.91KK75 pKa = 9.91DD76 pKa = 3.96TPLHH80 pKa = 5.84IKK82 pKa = 10.15KK83 pKa = 9.59LANYY87 pKa = 7.84FVSLRR92 pKa = 11.84VEE94 pKa = 4.07RR95 pKa = 11.84NFIDD99 pKa = 3.8SVIDD103 pKa = 3.1MSRR106 pKa = 11.84PKK108 pKa = 10.82LEE110 pKa = 5.1ADD112 pKa = 4.98DD113 pKa = 4.08IDD115 pKa = 4.07EE116 pKa = 4.55SFEE119 pKa = 4.12ALPTTGLSDD128 pKa = 3.49SKK130 pKa = 10.89KK131 pKa = 9.75IEE133 pKa = 4.18LEE135 pKa = 3.98QEE137 pKa = 4.09VQSILDD143 pKa = 4.33IINEE147 pKa = 4.14TDD149 pKa = 3.79VEE151 pKa = 4.2LQEE154 pKa = 4.66NAKK157 pKa = 10.4QYY159 pKa = 11.24EE160 pKa = 3.96KK161 pKa = 11.08DD162 pKa = 3.49LNLFEE167 pKa = 5.35EE168 pKa = 5.33DD169 pKa = 3.18IKK171 pKa = 11.31KK172 pKa = 10.51SGIEE176 pKa = 3.88IPPDD180 pKa = 3.39GLNADD185 pKa = 5.06FIEE188 pKa = 4.93TIQEE192 pKa = 4.09KK193 pKa = 10.79LSDD196 pKa = 3.58KK197 pKa = 10.82SQAILSQLIDD207 pKa = 3.42IAEE210 pKa = 4.31DD211 pKa = 3.56TLQKK215 pKa = 9.84KK216 pKa = 6.5TLKK219 pKa = 10.23EE220 pKa = 3.93EE221 pKa = 4.17KK222 pKa = 9.81KK223 pKa = 10.34QEE225 pKa = 3.73VDD227 pKa = 3.26ILIQQIEE234 pKa = 4.33EE235 pKa = 4.42SPVNIDD241 pKa = 3.88DD242 pKa = 3.95LKK244 pKa = 11.51NYY246 pKa = 10.02IFINRR251 pKa = 11.84STNEE255 pKa = 3.51FPYY258 pKa = 9.83YY259 pKa = 8.63TLEE262 pKa = 4.03YY263 pKa = 9.55KK264 pKa = 9.7KK265 pKa = 10.75QKK267 pKa = 9.81IDD269 pKa = 3.5EE270 pKa = 4.38LEE272 pKa = 4.14KK273 pKa = 8.93EE274 pKa = 4.22RR275 pKa = 11.84KK276 pKa = 9.7RR277 pKa = 11.84IGEE280 pKa = 4.17LIINEE285 pKa = 4.2VKK287 pKa = 10.69KK288 pKa = 11.11KK289 pKa = 10.21EE290 pKa = 3.92FF291 pKa = 3.7
MM1 pKa = 8.02PDD3 pKa = 3.55LVTPEE8 pKa = 3.91YY9 pKa = 10.89QNYY12 pKa = 7.56TFKK15 pKa = 11.02KK16 pKa = 9.57DD17 pKa = 3.66LIGAFTLEE25 pKa = 4.15EE26 pKa = 4.12LQSEE30 pKa = 4.43FQEE33 pKa = 3.99ISNEE37 pKa = 3.68FFRR40 pKa = 11.84IKK42 pKa = 10.51EE43 pKa = 4.0LIYY46 pKa = 11.06DD47 pKa = 3.87NLEE50 pKa = 3.93EE51 pKa = 4.3SPEE54 pKa = 3.98IVVPEE59 pKa = 4.38MGLEE63 pKa = 4.25DD64 pKa = 4.72PDD66 pKa = 5.64FIDD69 pKa = 4.43KK70 pKa = 10.69LVYY73 pKa = 10.83DD74 pKa = 4.91KK75 pKa = 9.91DD76 pKa = 3.96TPLHH80 pKa = 5.84IKK82 pKa = 10.15KK83 pKa = 9.59LANYY87 pKa = 7.84FVSLRR92 pKa = 11.84VEE94 pKa = 4.07RR95 pKa = 11.84NFIDD99 pKa = 3.8SVIDD103 pKa = 3.1MSRR106 pKa = 11.84PKK108 pKa = 10.82LEE110 pKa = 5.1ADD112 pKa = 4.98DD113 pKa = 4.08IDD115 pKa = 4.07EE116 pKa = 4.55SFEE119 pKa = 4.12ALPTTGLSDD128 pKa = 3.49SKK130 pKa = 10.89KK131 pKa = 9.75IEE133 pKa = 4.18LEE135 pKa = 3.98QEE137 pKa = 4.09VQSILDD143 pKa = 4.33IINEE147 pKa = 4.14TDD149 pKa = 3.79VEE151 pKa = 4.2LQEE154 pKa = 4.66NAKK157 pKa = 10.4QYY159 pKa = 11.24EE160 pKa = 3.96KK161 pKa = 11.08DD162 pKa = 3.49LNLFEE167 pKa = 5.35EE168 pKa = 5.33DD169 pKa = 3.18IKK171 pKa = 11.31KK172 pKa = 10.51SGIEE176 pKa = 3.88IPPDD180 pKa = 3.39GLNADD185 pKa = 5.06FIEE188 pKa = 4.93TIQEE192 pKa = 4.09KK193 pKa = 10.79LSDD196 pKa = 3.58KK197 pKa = 10.82SQAILSQLIDD207 pKa = 3.42IAEE210 pKa = 4.31DD211 pKa = 3.56TLQKK215 pKa = 9.84KK216 pKa = 6.5TLKK219 pKa = 10.23EE220 pKa = 3.93EE221 pKa = 4.17KK222 pKa = 9.81KK223 pKa = 10.34QEE225 pKa = 3.73VDD227 pKa = 3.26ILIQQIEE234 pKa = 4.33EE235 pKa = 4.42SPVNIDD241 pKa = 3.88DD242 pKa = 3.95LKK244 pKa = 11.51NYY246 pKa = 10.02IFINRR251 pKa = 11.84STNEE255 pKa = 3.51FPYY258 pKa = 9.83YY259 pKa = 8.63TLEE262 pKa = 4.03YY263 pKa = 9.55KK264 pKa = 9.7KK265 pKa = 10.75QKK267 pKa = 9.81IDD269 pKa = 3.5EE270 pKa = 4.38LEE272 pKa = 4.14KK273 pKa = 8.93EE274 pKa = 4.22RR275 pKa = 11.84KK276 pKa = 9.7RR277 pKa = 11.84IGEE280 pKa = 4.17LIINEE285 pKa = 4.2VKK287 pKa = 10.69KK288 pKa = 11.11KK289 pKa = 10.21EE290 pKa = 3.92FF291 pKa = 3.7
Molecular weight: 34.12 kDa
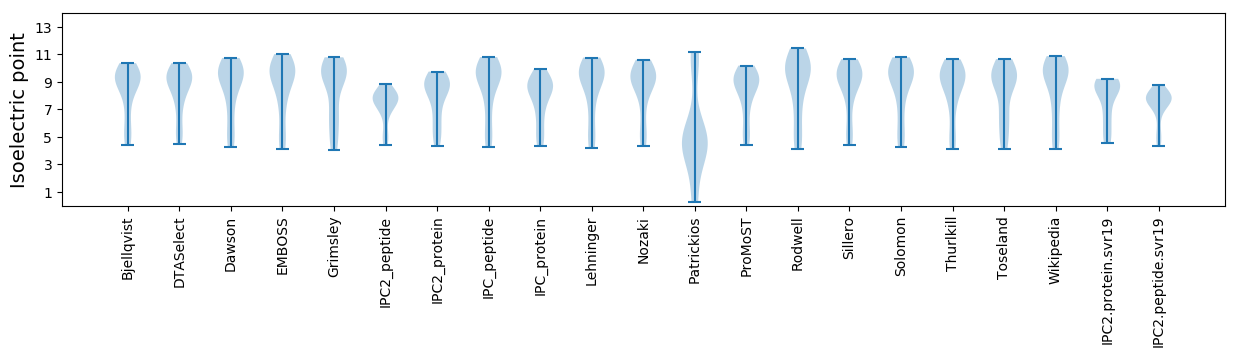
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A0RK92|A0A0A0RK92_9VIRU Uncharacterized protein OS=Yellowstone Lake virophage 6 OX=1557034 GN=YSLV6_ORF28 PE=4 SV=1
MM1 pKa = 7.1SAVRR5 pKa = 11.84GYY7 pKa = 11.2DD8 pKa = 3.14NLYY11 pKa = 10.0NEE13 pKa = 4.88HH14 pKa = 6.55VNDD17 pKa = 3.81IALMNYY23 pKa = 9.93NKK25 pKa = 10.61GIANRR30 pKa = 11.84LRR32 pKa = 11.84MQEE35 pKa = 3.51QAMQSKK41 pKa = 10.61LPFKK45 pKa = 10.44EE46 pKa = 3.93PQLLSGGVRR55 pKa = 11.84ASNSIVAGTTAEE67 pKa = 4.31PPSTLAVGGRR77 pKa = 11.84AFRR80 pKa = 11.84TFTGKK85 pKa = 7.74EE86 pKa = 4.1TPSQGGVNRR95 pKa = 11.84LKK97 pKa = 10.48KK98 pKa = 10.5AKK100 pKa = 10.15NGPHH104 pKa = 4.91TQKK107 pKa = 10.42IPWAMHH113 pKa = 5.38LTSQKK118 pKa = 8.72WRR120 pKa = 11.84VWVLKK125 pKa = 10.01VEE127 pKa = 4.01WFIVKK132 pKa = 9.79IYY134 pKa = 10.27KK135 pKa = 9.95KK136 pKa = 9.95HH137 pKa = 5.22
MM1 pKa = 7.1SAVRR5 pKa = 11.84GYY7 pKa = 11.2DD8 pKa = 3.14NLYY11 pKa = 10.0NEE13 pKa = 4.88HH14 pKa = 6.55VNDD17 pKa = 3.81IALMNYY23 pKa = 9.93NKK25 pKa = 10.61GIANRR30 pKa = 11.84LRR32 pKa = 11.84MQEE35 pKa = 3.51QAMQSKK41 pKa = 10.61LPFKK45 pKa = 10.44EE46 pKa = 3.93PQLLSGGVRR55 pKa = 11.84ASNSIVAGTTAEE67 pKa = 4.31PPSTLAVGGRR77 pKa = 11.84AFRR80 pKa = 11.84TFTGKK85 pKa = 7.74EE86 pKa = 4.1TPSQGGVNRR95 pKa = 11.84LKK97 pKa = 10.48KK98 pKa = 10.5AKK100 pKa = 10.15NGPHH104 pKa = 4.91TQKK107 pKa = 10.42IPWAMHH113 pKa = 5.38LTSQKK118 pKa = 8.72WRR120 pKa = 11.84VWVLKK125 pKa = 10.01VEE127 pKa = 4.01WFIVKK132 pKa = 9.79IYY134 pKa = 10.27KK135 pKa = 9.95KK136 pKa = 9.95HH137 pKa = 5.22
Molecular weight: 15.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7356 |
56 |
1070 |
253.7 |
28.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.568 ± 0.666 | 1.088 ± 0.232 |
5.166 ± 0.596 | 5.424 ± 0.968 |
3.806 ± 0.497 | 5.914 ± 0.998 |
0.843 ± 0.183 | 8.877 ± 0.344 |
9.122 ± 1.631 | 8.904 ± 0.353 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.672 ± 0.239 | 8.021 ± 0.339 |
3.399 ± 0.305 | 3.29 ± 0.263 |
3.263 ± 0.351 | 8.333 ± 1.015 |
7.817 ± 1.537 | 4.948 ± 0.315 |
0.816 ± 0.1 | 4.731 ± 0.395 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
