
Roseivirga spongicola
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Roseivirgaceae; Roseivirga
Average proteome isoelectric point is 6.09
Get precalculated fractions of proteins
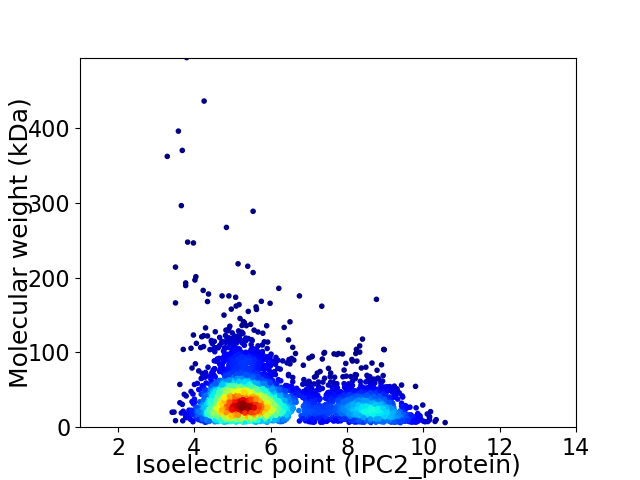
Virtual 2D-PAGE plot for 3854 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A150X5K0|A0A150X5K0_9BACT Phenylacetate-CoA oxygenase subunit PaaJ OS=Roseivirga spongicola OX=333140 GN=AWW68_15115 PE=4 SV=1
MM1 pKa = 8.0RR2 pKa = 11.84ILLVGCFILTCHH14 pKa = 6.21FVCAQGTFTSANSGSWTDD32 pKa = 3.47PGSWTLVGTDD42 pKa = 3.54SDD44 pKa = 4.18GNNYY48 pKa = 9.39PDD50 pKa = 4.61GNDD53 pKa = 2.99NVIINGHH60 pKa = 7.21AITVSSTEE68 pKa = 3.8ATSTLEE74 pKa = 3.82LATGIVEE81 pKa = 4.46FSGAAAQLNVRR92 pKa = 11.84GTLSSSGASEE102 pKa = 4.2VTGTSTNHH110 pKa = 6.48RR111 pKa = 11.84LVIVNNGTTVLGNISVNGGTFTLDD135 pKa = 3.41GARR138 pKa = 11.84LTNNGEE144 pKa = 3.93LTVADD149 pKa = 4.32GATFLIDD156 pKa = 4.76DD157 pKa = 4.06STAPTVVANHH167 pKa = 7.29LFTDD171 pKa = 3.66IDD173 pKa = 3.79INSNGTFQNNDD184 pKa = 3.48AIQIIIRR191 pKa = 11.84GTITNNGTFNGCGDD205 pKa = 3.95DD206 pKa = 3.94SCNYY210 pKa = 8.17TYY212 pKa = 11.32NVAGSFVVDD221 pKa = 3.89GTTTVNMPRR230 pKa = 11.84VLVYY234 pKa = 10.83NGATLTNQGLISTIDD249 pKa = 3.52LLGNGVGVNTFVNEE263 pKa = 3.76GTLTVNTSGTIDD275 pKa = 3.63EE276 pKa = 5.13ADD278 pKa = 3.42LVLDD282 pKa = 4.3FDD284 pKa = 4.83AVGNTLVYY292 pKa = 10.3EE293 pKa = 4.93GSTTEE298 pKa = 3.97QLFSSTFYY306 pKa = 10.57NVEE309 pKa = 3.93VNLEE313 pKa = 4.34DD314 pKa = 3.54GTLINLNNGVSSTINNNLTVTSGDD338 pKa = 3.18ARR340 pKa = 11.84IINSANTLTVLGNLSIEE357 pKa = 4.36GDD359 pKa = 3.37GSFFMNTTSVGEE371 pKa = 4.53GISITGDD378 pKa = 3.03FDD380 pKa = 4.22VSSTNATAVDD390 pKa = 4.7LNNGTITFTNMTITNGADD408 pKa = 3.08VFTGVADD415 pKa = 3.95LTSMGTITVTNGSVNPDD432 pKa = 2.85GTATPTFNNILVEE445 pKa = 4.78ANGSWLSTSVYY456 pKa = 10.87DD457 pKa = 3.83PTITGNITNNGTFTACLNTSGCDD480 pKa = 3.39YY481 pKa = 10.36TLTSGSGTLGGSSTISNMANIFLNDD506 pKa = 3.41GASYY510 pKa = 10.58TNEE513 pKa = 3.74NTSAGGIQVGTGIQTTSGAGSFINGANATLDD544 pKa = 3.68YY545 pKa = 11.18SGTLANFSVTNFTASAAGNTVIFSRR570 pKa = 11.84PDD572 pKa = 3.04ANQSLRR578 pKa = 11.84ATADD582 pKa = 3.18ANNEE586 pKa = 4.17YY587 pKa = 11.35VNVTINKK594 pKa = 9.51ADD596 pKa = 3.69GFDD599 pKa = 4.48VILDD603 pKa = 3.85DD604 pKa = 5.24NITVSGTLTLTAGDD618 pKa = 3.81VQLNAFDD625 pKa = 4.1LTLADD630 pKa = 3.98GATVSGGNSTSYY642 pKa = 10.8IKK644 pKa = 11.14LNGAGLLNQEE654 pKa = 4.27YY655 pKa = 9.76SAVGATLSFPIGDD668 pKa = 3.66VDD670 pKa = 5.19DD671 pKa = 5.07FSPINSLTLHH681 pKa = 6.31SATLGASPSIEE692 pKa = 4.19FDD694 pKa = 2.8ITDD697 pKa = 3.76AANSNRR703 pKa = 11.84DD704 pKa = 3.03VDD706 pKa = 3.85NTGAGGDD713 pKa = 4.12DD714 pKa = 4.96DD715 pKa = 4.61GTAATSFLSRR725 pKa = 11.84FWTLTPTDD733 pKa = 3.43ISDD736 pKa = 3.57VTFSASYY743 pKa = 9.29TYY745 pKa = 11.3VDD747 pKa = 4.08ADD749 pKa = 3.79VAGATANEE757 pKa = 4.09AEE759 pKa = 4.72LVGSVYY765 pKa = 9.29RR766 pKa = 11.84TPIGEE771 pKa = 4.52GFLDD775 pKa = 3.66WSVEE779 pKa = 4.03EE780 pKa = 4.54AVNATSNLVTVSNVSSTALGVLYY803 pKa = 11.06AMDD806 pKa = 3.78NTLEE810 pKa = 4.1RR811 pKa = 11.84LPVEE815 pKa = 4.28LLSFTAITRR824 pKa = 11.84LQEE827 pKa = 4.11VVLNWEE833 pKa = 4.38TTSEE837 pKa = 4.16EE838 pKa = 4.31NNSHH842 pKa = 5.06FTIEE846 pKa = 3.89RR847 pKa = 11.84SANGISFQSIGEE859 pKa = 4.13LEE861 pKa = 4.25GKK863 pKa = 10.34GNSSEE868 pKa = 4.38LISYY872 pKa = 9.31SFIDD876 pKa = 4.07EE877 pKa = 4.43FPLTGRR883 pKa = 11.84AYY885 pKa = 10.69YY886 pKa = 10.08RR887 pKa = 11.84LKK889 pKa = 10.71QVDD892 pKa = 3.77FSGAFEE898 pKa = 3.99YY899 pKa = 11.09SEE901 pKa = 4.74VISAVYY907 pKa = 10.36DD908 pKa = 3.46GAVNYY913 pKa = 9.95NFRR916 pKa = 11.84LFPNPVNSGGEE927 pKa = 3.93LTLSFEE933 pKa = 4.41RR934 pKa = 11.84TQTNSEE940 pKa = 4.23VVKK943 pKa = 9.76WQLLSITGEE952 pKa = 3.85QLIEE956 pKa = 3.91QVGSDD961 pKa = 3.59KK962 pKa = 11.03EE963 pKa = 4.26YY964 pKa = 10.43IIDD967 pKa = 3.74TSKK970 pKa = 11.25LSLGAYY976 pKa = 9.07LVRR979 pKa = 11.84VVYY982 pKa = 10.77SDD984 pKa = 3.71GRR986 pKa = 11.84VDD988 pKa = 3.61SQRR991 pKa = 11.84LVVNN995 pKa = 4.52
MM1 pKa = 8.0RR2 pKa = 11.84ILLVGCFILTCHH14 pKa = 6.21FVCAQGTFTSANSGSWTDD32 pKa = 3.47PGSWTLVGTDD42 pKa = 3.54SDD44 pKa = 4.18GNNYY48 pKa = 9.39PDD50 pKa = 4.61GNDD53 pKa = 2.99NVIINGHH60 pKa = 7.21AITVSSTEE68 pKa = 3.8ATSTLEE74 pKa = 3.82LATGIVEE81 pKa = 4.46FSGAAAQLNVRR92 pKa = 11.84GTLSSSGASEE102 pKa = 4.2VTGTSTNHH110 pKa = 6.48RR111 pKa = 11.84LVIVNNGTTVLGNISVNGGTFTLDD135 pKa = 3.41GARR138 pKa = 11.84LTNNGEE144 pKa = 3.93LTVADD149 pKa = 4.32GATFLIDD156 pKa = 4.76DD157 pKa = 4.06STAPTVVANHH167 pKa = 7.29LFTDD171 pKa = 3.66IDD173 pKa = 3.79INSNGTFQNNDD184 pKa = 3.48AIQIIIRR191 pKa = 11.84GTITNNGTFNGCGDD205 pKa = 3.95DD206 pKa = 3.94SCNYY210 pKa = 8.17TYY212 pKa = 11.32NVAGSFVVDD221 pKa = 3.89GTTTVNMPRR230 pKa = 11.84VLVYY234 pKa = 10.83NGATLTNQGLISTIDD249 pKa = 3.52LLGNGVGVNTFVNEE263 pKa = 3.76GTLTVNTSGTIDD275 pKa = 3.63EE276 pKa = 5.13ADD278 pKa = 3.42LVLDD282 pKa = 4.3FDD284 pKa = 4.83AVGNTLVYY292 pKa = 10.3EE293 pKa = 4.93GSTTEE298 pKa = 3.97QLFSSTFYY306 pKa = 10.57NVEE309 pKa = 3.93VNLEE313 pKa = 4.34DD314 pKa = 3.54GTLINLNNGVSSTINNNLTVTSGDD338 pKa = 3.18ARR340 pKa = 11.84IINSANTLTVLGNLSIEE357 pKa = 4.36GDD359 pKa = 3.37GSFFMNTTSVGEE371 pKa = 4.53GISITGDD378 pKa = 3.03FDD380 pKa = 4.22VSSTNATAVDD390 pKa = 4.7LNNGTITFTNMTITNGADD408 pKa = 3.08VFTGVADD415 pKa = 3.95LTSMGTITVTNGSVNPDD432 pKa = 2.85GTATPTFNNILVEE445 pKa = 4.78ANGSWLSTSVYY456 pKa = 10.87DD457 pKa = 3.83PTITGNITNNGTFTACLNTSGCDD480 pKa = 3.39YY481 pKa = 10.36TLTSGSGTLGGSSTISNMANIFLNDD506 pKa = 3.41GASYY510 pKa = 10.58TNEE513 pKa = 3.74NTSAGGIQVGTGIQTTSGAGSFINGANATLDD544 pKa = 3.68YY545 pKa = 11.18SGTLANFSVTNFTASAAGNTVIFSRR570 pKa = 11.84PDD572 pKa = 3.04ANQSLRR578 pKa = 11.84ATADD582 pKa = 3.18ANNEE586 pKa = 4.17YY587 pKa = 11.35VNVTINKK594 pKa = 9.51ADD596 pKa = 3.69GFDD599 pKa = 4.48VILDD603 pKa = 3.85DD604 pKa = 5.24NITVSGTLTLTAGDD618 pKa = 3.81VQLNAFDD625 pKa = 4.1LTLADD630 pKa = 3.98GATVSGGNSTSYY642 pKa = 10.8IKK644 pKa = 11.14LNGAGLLNQEE654 pKa = 4.27YY655 pKa = 9.76SAVGATLSFPIGDD668 pKa = 3.66VDD670 pKa = 5.19DD671 pKa = 5.07FSPINSLTLHH681 pKa = 6.31SATLGASPSIEE692 pKa = 4.19FDD694 pKa = 2.8ITDD697 pKa = 3.76AANSNRR703 pKa = 11.84DD704 pKa = 3.03VDD706 pKa = 3.85NTGAGGDD713 pKa = 4.12DD714 pKa = 4.96DD715 pKa = 4.61GTAATSFLSRR725 pKa = 11.84FWTLTPTDD733 pKa = 3.43ISDD736 pKa = 3.57VTFSASYY743 pKa = 9.29TYY745 pKa = 11.3VDD747 pKa = 4.08ADD749 pKa = 3.79VAGATANEE757 pKa = 4.09AEE759 pKa = 4.72LVGSVYY765 pKa = 9.29RR766 pKa = 11.84TPIGEE771 pKa = 4.52GFLDD775 pKa = 3.66WSVEE779 pKa = 4.03EE780 pKa = 4.54AVNATSNLVTVSNVSSTALGVLYY803 pKa = 11.06AMDD806 pKa = 3.78NTLEE810 pKa = 4.1RR811 pKa = 11.84LPVEE815 pKa = 4.28LLSFTAITRR824 pKa = 11.84LQEE827 pKa = 4.11VVLNWEE833 pKa = 4.38TTSEE837 pKa = 4.16EE838 pKa = 4.31NNSHH842 pKa = 5.06FTIEE846 pKa = 3.89RR847 pKa = 11.84SANGISFQSIGEE859 pKa = 4.13LEE861 pKa = 4.25GKK863 pKa = 10.34GNSSEE868 pKa = 4.38LISYY872 pKa = 9.31SFIDD876 pKa = 4.07EE877 pKa = 4.43FPLTGRR883 pKa = 11.84AYY885 pKa = 10.69YY886 pKa = 10.08RR887 pKa = 11.84LKK889 pKa = 10.71QVDD892 pKa = 3.77FSGAFEE898 pKa = 3.99YY899 pKa = 11.09SEE901 pKa = 4.74VISAVYY907 pKa = 10.36DD908 pKa = 3.46GAVNYY913 pKa = 9.95NFRR916 pKa = 11.84LFPNPVNSGGEE927 pKa = 3.93LTLSFEE933 pKa = 4.41RR934 pKa = 11.84TQTNSEE940 pKa = 4.23VVKK943 pKa = 9.76WQLLSITGEE952 pKa = 3.85QLIEE956 pKa = 3.91QVGSDD961 pKa = 3.59KK962 pKa = 11.03EE963 pKa = 4.26YY964 pKa = 10.43IIDD967 pKa = 3.74TSKK970 pKa = 11.25LSLGAYY976 pKa = 9.07LVRR979 pKa = 11.84VVYY982 pKa = 10.77SDD984 pKa = 3.71GRR986 pKa = 11.84VDD988 pKa = 3.61SQRR991 pKa = 11.84LVVNN995 pKa = 4.52
Molecular weight: 103.99 kDa
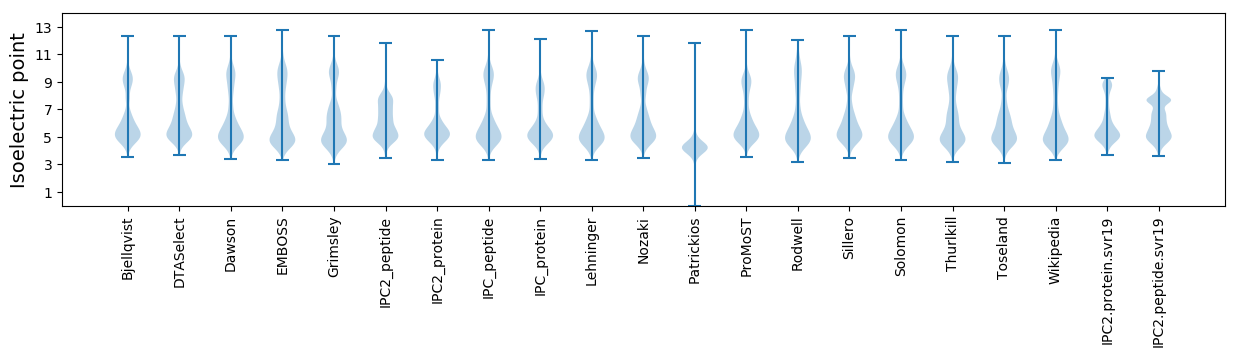
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A150XBP5|A0A150XBP5_9BACT Uncharacterized protein OS=Roseivirga spongicola OX=333140 GN=AWW68_10030 PE=4 SV=1
MM1 pKa = 7.52RR2 pKa = 11.84TTLLSNSIFLITSSFLRR19 pKa = 11.84ISILQKK25 pKa = 9.37YY26 pKa = 9.41RR27 pKa = 11.84GRR29 pKa = 11.84KK30 pKa = 7.84HH31 pKa = 6.25VKK33 pKa = 9.65SEE35 pKa = 3.73NYY37 pKa = 9.46QIRR40 pKa = 11.84EE41 pKa = 4.3LNSSDD46 pKa = 4.1LKK48 pKa = 11.01HH49 pKa = 6.92SNVDD53 pKa = 3.72FWTIAGRR60 pKa = 11.84IAIAAGLLRR69 pKa = 11.84AAEE72 pKa = 4.28DD73 pKa = 3.5VFLRR77 pKa = 11.84GTT79 pKa = 3.46
MM1 pKa = 7.52RR2 pKa = 11.84TTLLSNSIFLITSSFLRR19 pKa = 11.84ISILQKK25 pKa = 9.37YY26 pKa = 9.41RR27 pKa = 11.84GRR29 pKa = 11.84KK30 pKa = 7.84HH31 pKa = 6.25VKK33 pKa = 9.65SEE35 pKa = 3.73NYY37 pKa = 9.46QIRR40 pKa = 11.84EE41 pKa = 4.3LNSSDD46 pKa = 4.1LKK48 pKa = 11.01HH49 pKa = 6.92SNVDD53 pKa = 3.72FWTIAGRR60 pKa = 11.84IAIAAGLLRR69 pKa = 11.84AAEE72 pKa = 4.28DD73 pKa = 3.5VFLRR77 pKa = 11.84GTT79 pKa = 3.46
Molecular weight: 9.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1330867 |
52 |
4624 |
345.3 |
38.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.798 ± 0.041 | 0.654 ± 0.011 |
5.621 ± 0.034 | 7.216 ± 0.044 |
5.09 ± 0.03 | 6.807 ± 0.044 |
1.742 ± 0.02 | 7.104 ± 0.031 |
6.743 ± 0.058 | 9.543 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.363 ± 0.025 | 5.395 ± 0.04 |
3.514 ± 0.021 | 3.712 ± 0.022 |
4.029 ± 0.031 | 6.828 ± 0.04 |
5.372 ± 0.064 | 6.499 ± 0.034 |
1.147 ± 0.015 | 3.824 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
