
Cucurbit aphid borne yellows virus associated RNA
Taxonomy: Viruses; Riboviria; Virus-associated RNAs
Average proteome isoelectric point is 8.55
Get precalculated fractions of proteins
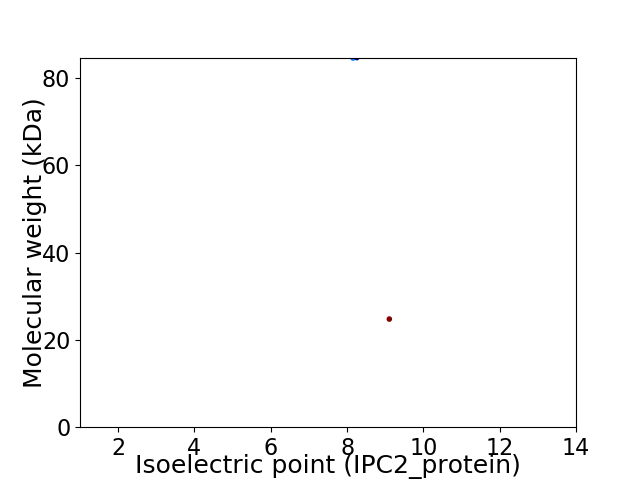
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5BKM2|A0A0C5BKM2_9VIRU RNA-directed RNA polymerase OS=Cucurbit aphid borne yellows virus associated RNA OX=1611875 PE=4 SV=1
MM1 pKa = 7.66KK2 pKa = 9.39MQLPSFSVGQHH13 pKa = 4.46ITSKK17 pKa = 10.76LVGVMPKK24 pKa = 10.33LGNKK28 pKa = 9.15LATAGKK34 pKa = 10.21VSATCGLVVAAGFSLYY50 pKa = 10.38AYY52 pKa = 10.21RR53 pKa = 11.84KK54 pKa = 8.29VCINKK59 pKa = 10.02LEE61 pKa = 4.09LVRR64 pKa = 11.84INTVPLPDD72 pKa = 4.57DD73 pKa = 4.64CIEE76 pKa = 4.8DD77 pKa = 3.81ALEE80 pKa = 4.38MEE82 pKa = 5.56DD83 pKa = 4.45DD84 pKa = 3.89AASEE88 pKa = 4.03LVQAKK93 pKa = 9.14IAKK96 pKa = 8.27EE97 pKa = 3.73AAIAAALEE105 pKa = 4.18PTKK108 pKa = 10.28PAEE111 pKa = 4.14QFAAAAKK118 pKa = 9.92IRR120 pKa = 11.84AQPPPNPLAKK130 pKa = 10.2RR131 pKa = 11.84MNRR134 pKa = 11.84LRR136 pKa = 11.84PGSTGKK142 pKa = 10.63FIRR145 pKa = 11.84VVRR148 pKa = 11.84AEE150 pKa = 4.4IKK152 pKa = 10.6AQMGTPTITAANEE165 pKa = 3.47AVIRR169 pKa = 11.84HH170 pKa = 5.67MLSKK174 pKa = 10.67YY175 pKa = 9.77CAVRR179 pKa = 11.84NIRR182 pKa = 11.84TSSYY186 pKa = 8.16AHH188 pKa = 5.96LVSRR192 pKa = 11.84VVRR195 pKa = 11.84EE196 pKa = 3.73VMTPYY201 pKa = 10.41PGDD204 pKa = 3.58SEE206 pKa = 4.35QVEE209 pKa = 4.44RR210 pKa = 11.84ASSVVNRR217 pKa = 11.84IHH219 pKa = 6.44NWLVQYY225 pKa = 10.67KK226 pKa = 9.79KK227 pKa = 8.89XGGLEE232 pKa = 3.81LARR235 pKa = 11.84GFTHH239 pKa = 7.0NEE241 pKa = 3.32TRR243 pKa = 11.84TDD245 pKa = 3.64VPGLDD250 pKa = 3.31ATNLGEE256 pKa = 4.81SRR258 pKa = 11.84PLTDD262 pKa = 3.02TNIRR266 pKa = 11.84RR267 pKa = 11.84VVGPVSSDD275 pKa = 3.14YY276 pKa = 11.43DD277 pKa = 3.33IVFYY281 pKa = 10.91TNSRR285 pKa = 11.84NNLMRR290 pKa = 11.84GLVNRR295 pKa = 11.84VLTYY299 pKa = 10.52KK300 pKa = 10.51GGPVLEE306 pKa = 4.38PTPGAWRR313 pKa = 11.84EE314 pKa = 3.99LRR316 pKa = 11.84SLAHH320 pKa = 6.52SLGNRR325 pKa = 11.84CLTTPLSSEE334 pKa = 4.22EE335 pKa = 4.09FLACYY340 pKa = 9.93AGRR343 pKa = 11.84KK344 pKa = 7.38RR345 pKa = 11.84AIYY348 pKa = 10.04AKK350 pKa = 10.25AIEE353 pKa = 4.46SFNTKK358 pKa = 9.49PWDD361 pKa = 3.56ADD363 pKa = 2.67KK364 pKa = 11.83DD365 pKa = 4.06MIVKK369 pKa = 10.42AFIKK373 pKa = 10.43KK374 pKa = 10.09EE375 pKa = 3.78KK376 pKa = 10.62DD377 pKa = 2.99KK378 pKa = 11.24LLSSNSDD385 pKa = 3.05PRR387 pKa = 11.84IIQPRR392 pKa = 11.84HH393 pKa = 4.93PRR395 pKa = 11.84FLVSLGRR402 pKa = 11.84YY403 pKa = 8.4IRR405 pKa = 11.84PLEE408 pKa = 3.8QKK410 pKa = 10.11VYY412 pKa = 9.13KK413 pKa = 10.2TYY415 pKa = 10.28TRR417 pKa = 11.84LFKK420 pKa = 10.79RR421 pKa = 11.84FAATNTPVCFKK432 pKa = 10.51GLNYY436 pKa = 9.99LRR438 pKa = 11.84RR439 pKa = 11.84GSCLKK444 pKa = 10.55EE445 pKa = 3.51KK446 pKa = 10.09WEE448 pKa = 4.43SFNSPVAVCLDD459 pKa = 3.62ASRR462 pKa = 11.84FDD464 pKa = 3.57LHH466 pKa = 8.19VSEE469 pKa = 6.09DD470 pKa = 4.03ALQHH474 pKa = 4.57THH476 pKa = 6.44LLYY479 pKa = 9.86RR480 pKa = 11.84TAIGNDD486 pKa = 3.4PLLAYY491 pKa = 9.57ILEE494 pKa = 4.39NRR496 pKa = 11.84LVTDD500 pKa = 4.44GVGWSSDD507 pKa = 2.88GAYY510 pKa = 9.8RR511 pKa = 11.84YY512 pKa = 9.02HH513 pKa = 7.04KK514 pKa = 10.61RR515 pKa = 11.84GGRR518 pKa = 11.84CSGDD522 pKa = 3.04NDD524 pKa = 3.61TSLGNVIIMLSITHH538 pKa = 6.95AFCEE542 pKa = 4.37EE543 pKa = 4.14SEE545 pKa = 4.21IPHH548 pKa = 7.15IEE550 pKa = 4.07VANDD554 pKa = 3.32GDD556 pKa = 3.9DD557 pKa = 3.24QVIIVEE563 pKa = 4.37SEE565 pKa = 3.95HH566 pKa = 6.94VEE568 pKa = 3.82KK569 pKa = 10.87VLRR572 pKa = 11.84IEE574 pKa = 5.12DD575 pKa = 3.37IFRR578 pKa = 11.84EE579 pKa = 4.24FGFLLKK585 pKa = 10.6VEE587 pKa = 4.53EE588 pKa = 4.52PVHH591 pKa = 5.09EE592 pKa = 4.32LEE594 pKa = 6.34RR595 pKa = 11.84IDD597 pKa = 4.2FCQTRR602 pKa = 11.84PVQLTPDD609 pKa = 3.24VCTMVRR615 pKa = 11.84HH616 pKa = 6.23PRR618 pKa = 11.84LAMTKK623 pKa = 10.27DD624 pKa = 3.25LTTFIPIEE632 pKa = 4.15RR633 pKa = 11.84GKK635 pKa = 10.76LKK637 pKa = 10.85YY638 pKa = 10.34HH639 pKa = 6.45MLTAIGKK646 pKa = 9.8CGMACYY652 pKa = 10.66NDD654 pKa = 3.76IPVLGAMYY662 pKa = 10.26RR663 pKa = 11.84KK664 pKa = 9.18LVKK667 pKa = 9.92IGSMNKK673 pKa = 9.81GNEE676 pKa = 3.85EE677 pKa = 4.01RR678 pKa = 11.84WWKK681 pKa = 8.27STHH684 pKa = 6.77ADD686 pKa = 3.3PAFRR690 pKa = 11.84SLSSKK695 pKa = 10.51AATNVGLTPQCRR707 pKa = 11.84ASFWKK712 pKa = 10.72AFGILPDD719 pKa = 3.57QQVALEE725 pKa = 4.51KK726 pKa = 10.32EE727 pKa = 4.31WNDD730 pKa = 3.53WEE732 pKa = 4.24PLLDD736 pKa = 4.61KK737 pKa = 10.78IEE739 pKa = 4.26YY740 pKa = 8.68VHH742 pKa = 6.81SFLGDD747 pKa = 3.54YY748 pKa = 7.89PVCEE752 pKa = 4.23GDD754 pKa = 3.51VV755 pKa = 3.25
MM1 pKa = 7.66KK2 pKa = 9.39MQLPSFSVGQHH13 pKa = 4.46ITSKK17 pKa = 10.76LVGVMPKK24 pKa = 10.33LGNKK28 pKa = 9.15LATAGKK34 pKa = 10.21VSATCGLVVAAGFSLYY50 pKa = 10.38AYY52 pKa = 10.21RR53 pKa = 11.84KK54 pKa = 8.29VCINKK59 pKa = 10.02LEE61 pKa = 4.09LVRR64 pKa = 11.84INTVPLPDD72 pKa = 4.57DD73 pKa = 4.64CIEE76 pKa = 4.8DD77 pKa = 3.81ALEE80 pKa = 4.38MEE82 pKa = 5.56DD83 pKa = 4.45DD84 pKa = 3.89AASEE88 pKa = 4.03LVQAKK93 pKa = 9.14IAKK96 pKa = 8.27EE97 pKa = 3.73AAIAAALEE105 pKa = 4.18PTKK108 pKa = 10.28PAEE111 pKa = 4.14QFAAAAKK118 pKa = 9.92IRR120 pKa = 11.84AQPPPNPLAKK130 pKa = 10.2RR131 pKa = 11.84MNRR134 pKa = 11.84LRR136 pKa = 11.84PGSTGKK142 pKa = 10.63FIRR145 pKa = 11.84VVRR148 pKa = 11.84AEE150 pKa = 4.4IKK152 pKa = 10.6AQMGTPTITAANEE165 pKa = 3.47AVIRR169 pKa = 11.84HH170 pKa = 5.67MLSKK174 pKa = 10.67YY175 pKa = 9.77CAVRR179 pKa = 11.84NIRR182 pKa = 11.84TSSYY186 pKa = 8.16AHH188 pKa = 5.96LVSRR192 pKa = 11.84VVRR195 pKa = 11.84EE196 pKa = 3.73VMTPYY201 pKa = 10.41PGDD204 pKa = 3.58SEE206 pKa = 4.35QVEE209 pKa = 4.44RR210 pKa = 11.84ASSVVNRR217 pKa = 11.84IHH219 pKa = 6.44NWLVQYY225 pKa = 10.67KK226 pKa = 9.79KK227 pKa = 8.89XGGLEE232 pKa = 3.81LARR235 pKa = 11.84GFTHH239 pKa = 7.0NEE241 pKa = 3.32TRR243 pKa = 11.84TDD245 pKa = 3.64VPGLDD250 pKa = 3.31ATNLGEE256 pKa = 4.81SRR258 pKa = 11.84PLTDD262 pKa = 3.02TNIRR266 pKa = 11.84RR267 pKa = 11.84VVGPVSSDD275 pKa = 3.14YY276 pKa = 11.43DD277 pKa = 3.33IVFYY281 pKa = 10.91TNSRR285 pKa = 11.84NNLMRR290 pKa = 11.84GLVNRR295 pKa = 11.84VLTYY299 pKa = 10.52KK300 pKa = 10.51GGPVLEE306 pKa = 4.38PTPGAWRR313 pKa = 11.84EE314 pKa = 3.99LRR316 pKa = 11.84SLAHH320 pKa = 6.52SLGNRR325 pKa = 11.84CLTTPLSSEE334 pKa = 4.22EE335 pKa = 4.09FLACYY340 pKa = 9.93AGRR343 pKa = 11.84KK344 pKa = 7.38RR345 pKa = 11.84AIYY348 pKa = 10.04AKK350 pKa = 10.25AIEE353 pKa = 4.46SFNTKK358 pKa = 9.49PWDD361 pKa = 3.56ADD363 pKa = 2.67KK364 pKa = 11.83DD365 pKa = 4.06MIVKK369 pKa = 10.42AFIKK373 pKa = 10.43KK374 pKa = 10.09EE375 pKa = 3.78KK376 pKa = 10.62DD377 pKa = 2.99KK378 pKa = 11.24LLSSNSDD385 pKa = 3.05PRR387 pKa = 11.84IIQPRR392 pKa = 11.84HH393 pKa = 4.93PRR395 pKa = 11.84FLVSLGRR402 pKa = 11.84YY403 pKa = 8.4IRR405 pKa = 11.84PLEE408 pKa = 3.8QKK410 pKa = 10.11VYY412 pKa = 9.13KK413 pKa = 10.2TYY415 pKa = 10.28TRR417 pKa = 11.84LFKK420 pKa = 10.79RR421 pKa = 11.84FAATNTPVCFKK432 pKa = 10.51GLNYY436 pKa = 9.99LRR438 pKa = 11.84RR439 pKa = 11.84GSCLKK444 pKa = 10.55EE445 pKa = 3.51KK446 pKa = 10.09WEE448 pKa = 4.43SFNSPVAVCLDD459 pKa = 3.62ASRR462 pKa = 11.84FDD464 pKa = 3.57LHH466 pKa = 8.19VSEE469 pKa = 6.09DD470 pKa = 4.03ALQHH474 pKa = 4.57THH476 pKa = 6.44LLYY479 pKa = 9.86RR480 pKa = 11.84TAIGNDD486 pKa = 3.4PLLAYY491 pKa = 9.57ILEE494 pKa = 4.39NRR496 pKa = 11.84LVTDD500 pKa = 4.44GVGWSSDD507 pKa = 2.88GAYY510 pKa = 9.8RR511 pKa = 11.84YY512 pKa = 9.02HH513 pKa = 7.04KK514 pKa = 10.61RR515 pKa = 11.84GGRR518 pKa = 11.84CSGDD522 pKa = 3.04NDD524 pKa = 3.61TSLGNVIIMLSITHH538 pKa = 6.95AFCEE542 pKa = 4.37EE543 pKa = 4.14SEE545 pKa = 4.21IPHH548 pKa = 7.15IEE550 pKa = 4.07VANDD554 pKa = 3.32GDD556 pKa = 3.9DD557 pKa = 3.24QVIIVEE563 pKa = 4.37SEE565 pKa = 3.95HH566 pKa = 6.94VEE568 pKa = 3.82KK569 pKa = 10.87VLRR572 pKa = 11.84IEE574 pKa = 5.12DD575 pKa = 3.37IFRR578 pKa = 11.84EE579 pKa = 4.24FGFLLKK585 pKa = 10.6VEE587 pKa = 4.53EE588 pKa = 4.52PVHH591 pKa = 5.09EE592 pKa = 4.32LEE594 pKa = 6.34RR595 pKa = 11.84IDD597 pKa = 4.2FCQTRR602 pKa = 11.84PVQLTPDD609 pKa = 3.24VCTMVRR615 pKa = 11.84HH616 pKa = 6.23PRR618 pKa = 11.84LAMTKK623 pKa = 10.27DD624 pKa = 3.25LTTFIPIEE632 pKa = 4.15RR633 pKa = 11.84GKK635 pKa = 10.76LKK637 pKa = 10.85YY638 pKa = 10.34HH639 pKa = 6.45MLTAIGKK646 pKa = 9.8CGMACYY652 pKa = 10.66NDD654 pKa = 3.76IPVLGAMYY662 pKa = 10.26RR663 pKa = 11.84KK664 pKa = 9.18LVKK667 pKa = 9.92IGSMNKK673 pKa = 9.81GNEE676 pKa = 3.85EE677 pKa = 4.01RR678 pKa = 11.84WWKK681 pKa = 8.27STHH684 pKa = 6.77ADD686 pKa = 3.3PAFRR690 pKa = 11.84SLSSKK695 pKa = 10.51AATNVGLTPQCRR707 pKa = 11.84ASFWKK712 pKa = 10.72AFGILPDD719 pKa = 3.57QQVALEE725 pKa = 4.51KK726 pKa = 10.32EE727 pKa = 4.31WNDD730 pKa = 3.53WEE732 pKa = 4.24PLLDD736 pKa = 4.61KK737 pKa = 10.78IEE739 pKa = 4.26YY740 pKa = 8.68VHH742 pKa = 6.81SFLGDD747 pKa = 3.54YY748 pKa = 7.89PVCEE752 pKa = 4.23GDD754 pKa = 3.51VV755 pKa = 3.25
Molecular weight: 84.54 kDa
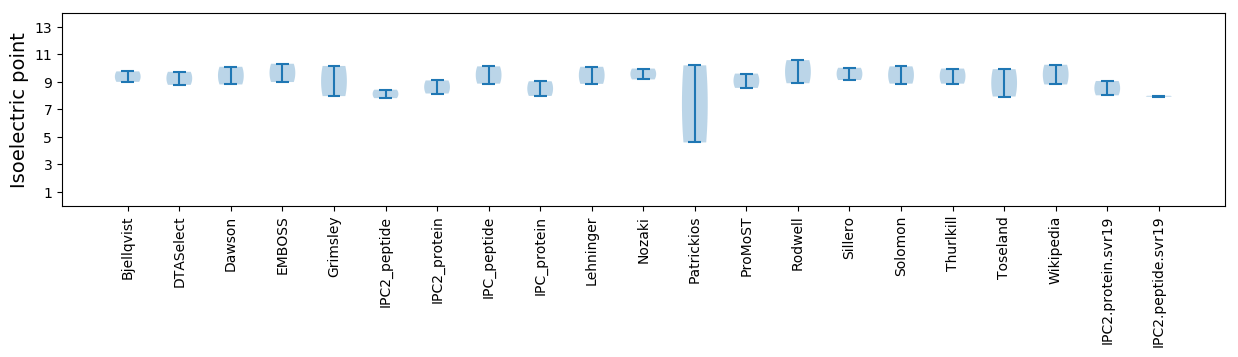
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5BKM2|A0A0C5BKM2_9VIRU RNA-directed RNA polymerase OS=Cucurbit aphid borne yellows virus associated RNA OX=1611875 PE=4 SV=1
MM1 pKa = 7.66KK2 pKa = 9.39MQLPSFSVGQHH13 pKa = 4.46ITSKK17 pKa = 10.76LVGVMPKK24 pKa = 10.33LGNKK28 pKa = 9.15LATAGKK34 pKa = 10.21VSATCGLVVAAGFSLYY50 pKa = 10.38AYY52 pKa = 10.21RR53 pKa = 11.84KK54 pKa = 8.29VCINKK59 pKa = 10.02LEE61 pKa = 4.09LVRR64 pKa = 11.84INTVPLPDD72 pKa = 4.57DD73 pKa = 4.64CIEE76 pKa = 4.8DD77 pKa = 3.81ALEE80 pKa = 4.38MEE82 pKa = 5.56DD83 pKa = 4.45DD84 pKa = 3.89AASEE88 pKa = 4.03LVQAKK93 pKa = 9.14IAKK96 pKa = 8.27EE97 pKa = 3.73AAIAAALEE105 pKa = 4.18PTKK108 pKa = 10.28PAEE111 pKa = 4.14QFAAAAKK118 pKa = 9.92IRR120 pKa = 11.84AQPPPNPLAKK130 pKa = 10.2RR131 pKa = 11.84MNRR134 pKa = 11.84LRR136 pKa = 11.84PGSTGKK142 pKa = 10.63FIRR145 pKa = 11.84VVRR148 pKa = 11.84AEE150 pKa = 4.4IKK152 pKa = 10.6AQMGTPTITAANEE165 pKa = 3.47AVIRR169 pKa = 11.84HH170 pKa = 5.67MLSKK174 pKa = 10.67YY175 pKa = 9.77CAVRR179 pKa = 11.84NIRR182 pKa = 11.84TSSYY186 pKa = 8.16AHH188 pKa = 5.96LVSRR192 pKa = 11.84VVRR195 pKa = 11.84EE196 pKa = 3.73VMTPYY201 pKa = 10.41PGDD204 pKa = 3.58SEE206 pKa = 4.35QVEE209 pKa = 4.44RR210 pKa = 11.84ASSVVNRR217 pKa = 11.84IHH219 pKa = 6.44NWLVQYY225 pKa = 10.87KK226 pKa = 9.86KK227 pKa = 10.91
MM1 pKa = 7.66KK2 pKa = 9.39MQLPSFSVGQHH13 pKa = 4.46ITSKK17 pKa = 10.76LVGVMPKK24 pKa = 10.33LGNKK28 pKa = 9.15LATAGKK34 pKa = 10.21VSATCGLVVAAGFSLYY50 pKa = 10.38AYY52 pKa = 10.21RR53 pKa = 11.84KK54 pKa = 8.29VCINKK59 pKa = 10.02LEE61 pKa = 4.09LVRR64 pKa = 11.84INTVPLPDD72 pKa = 4.57DD73 pKa = 4.64CIEE76 pKa = 4.8DD77 pKa = 3.81ALEE80 pKa = 4.38MEE82 pKa = 5.56DD83 pKa = 4.45DD84 pKa = 3.89AASEE88 pKa = 4.03LVQAKK93 pKa = 9.14IAKK96 pKa = 8.27EE97 pKa = 3.73AAIAAALEE105 pKa = 4.18PTKK108 pKa = 10.28PAEE111 pKa = 4.14QFAAAAKK118 pKa = 9.92IRR120 pKa = 11.84AQPPPNPLAKK130 pKa = 10.2RR131 pKa = 11.84MNRR134 pKa = 11.84LRR136 pKa = 11.84PGSTGKK142 pKa = 10.63FIRR145 pKa = 11.84VVRR148 pKa = 11.84AEE150 pKa = 4.4IKK152 pKa = 10.6AQMGTPTITAANEE165 pKa = 3.47AVIRR169 pKa = 11.84HH170 pKa = 5.67MLSKK174 pKa = 10.67YY175 pKa = 9.77CAVRR179 pKa = 11.84NIRR182 pKa = 11.84TSSYY186 pKa = 8.16AHH188 pKa = 5.96LVSRR192 pKa = 11.84VVRR195 pKa = 11.84EE196 pKa = 3.73VMTPYY201 pKa = 10.41PGDD204 pKa = 3.58SEE206 pKa = 4.35QVEE209 pKa = 4.44RR210 pKa = 11.84ASSVVNRR217 pKa = 11.84IHH219 pKa = 6.44NWLVQYY225 pKa = 10.87KK226 pKa = 9.86KK227 pKa = 10.91
Molecular weight: 24.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
982 |
227 |
755 |
491.0 |
54.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.572 ± 1.908 | 2.138 ± 0.176 |
4.481 ± 0.858 | 6.212 ± 0.227 |
2.953 ± 0.556 | 5.601 ± 0.558 |
2.342 ± 0.271 | 5.499 ± 0.106 |
6.619 ± 0.406 | 8.859 ± 0.64 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.546 ± 0.457 | 4.175 ± 0.098 |
5.601 ± 0.265 | 2.546 ± 0.457 |
7.026 ± 0.195 | 6.11 ± 0.027 |
5.397 ± 0.258 | 8.045 ± 0.769 |
1.12 ± 0.317 | 3.055 ± 0.192 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
