
Wenling tombus-like virus 4
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
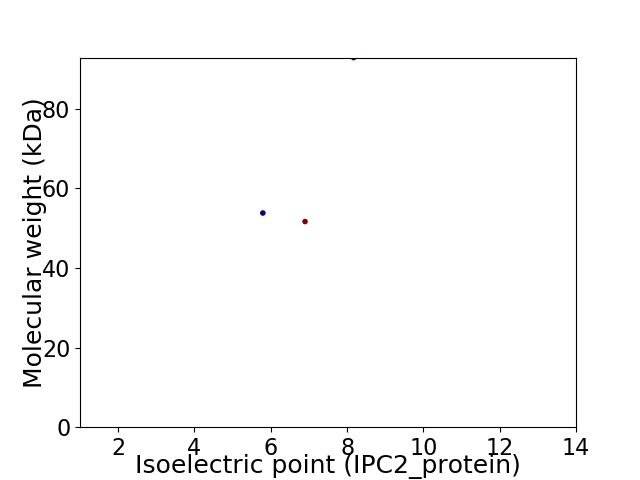
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KH34|A0A1L3KH34_9VIRU RNA-directed RNA polymerase OS=Wenling tombus-like virus 4 OX=1923546 PE=4 SV=1
MM1 pKa = 7.84PCMEE5 pKa = 5.44DD6 pKa = 2.8VTMATTVEE14 pKa = 4.24ATDD17 pKa = 3.4SSIWARR23 pKa = 11.84KK24 pKa = 8.33VPHH27 pKa = 6.19SVAIATAEE35 pKa = 4.08QVCLLEE41 pKa = 4.29TVEE44 pKa = 4.32IRR46 pKa = 11.84VPHH49 pKa = 6.7PLHH52 pKa = 6.58PGKK55 pKa = 10.81GLTDD59 pKa = 4.96LNTLAAQFLASKK71 pKa = 9.35PNQWLTQVEE80 pKa = 4.41PTPFQTWVKK89 pKa = 9.52RR90 pKa = 11.84FSPSRR95 pKa = 11.84QTQLEE100 pKa = 3.95AGRR103 pKa = 11.84EE104 pKa = 3.84WVARR108 pKa = 11.84HH109 pKa = 5.68GLLEE113 pKa = 4.65KK114 pKa = 10.7DD115 pKa = 3.44SIMAVFVKK123 pKa = 10.78VEE125 pKa = 4.04TSVTGTDD132 pKa = 3.66PRR134 pKa = 11.84NISPRR139 pKa = 11.84DD140 pKa = 3.41PRR142 pKa = 11.84FLSTLGPFTSRR153 pKa = 11.84LEE155 pKa = 4.02QLAVSCPYY163 pKa = 10.08LVKK166 pKa = 10.88GLDD169 pKa = 3.34TYY171 pKa = 11.6KK172 pKa = 10.57RR173 pKa = 11.84GPLLASYY180 pKa = 8.96WRR182 pKa = 11.84ACVIEE187 pKa = 4.16TDD189 pKa = 4.45FSRR192 pKa = 11.84FDD194 pKa = 3.36MTVSQDD200 pKa = 3.12IINTVEE206 pKa = 3.6RR207 pKa = 11.84SLFRR211 pKa = 11.84RR212 pKa = 11.84CFPEE216 pKa = 4.46GLYY219 pKa = 10.14PDD221 pKa = 5.62LDD223 pKa = 3.98ACLSYY228 pKa = 11.02LDD230 pKa = 4.17TMTGVSDD237 pKa = 3.96LGIMYY242 pKa = 7.69TVSGTRR248 pKa = 11.84ASGDD252 pKa = 3.06AHH254 pKa = 5.77TSIANGYY261 pKa = 8.78INRR264 pKa = 11.84FVIWSCLRR272 pKa = 11.84HH273 pKa = 6.35LDD275 pKa = 3.54PSKK278 pKa = 10.22WDD280 pKa = 3.65SFHH283 pKa = 7.05EE284 pKa = 4.71GDD286 pKa = 5.06DD287 pKa = 3.8GVIFLDD293 pKa = 3.38QDD295 pKa = 3.13IFEE298 pKa = 4.6TARR301 pKa = 11.84EE302 pKa = 4.04CLKK305 pKa = 10.64FASCLGFKK313 pKa = 10.63LKK315 pKa = 10.51VICPEE320 pKa = 4.17SQDD323 pKa = 3.47SAVFCGRR330 pKa = 11.84FTCTTCHH337 pKa = 7.09RR338 pKa = 11.84EE339 pKa = 4.04YY340 pKa = 11.05CDD342 pKa = 4.52LPRR345 pKa = 11.84ALSKK349 pKa = 10.67FHH351 pKa = 6.19TTTKK355 pKa = 10.93NGDD358 pKa = 3.78CKK360 pKa = 11.25SLVLAKK366 pKa = 10.35ALSYY370 pKa = 10.38FATDD374 pKa = 2.36AHH376 pKa = 5.73TPIMGVLCDD385 pKa = 4.56ALIKK389 pKa = 10.62LLSPLVSQRR398 pKa = 11.84LMRR401 pKa = 11.84RR402 pKa = 11.84RR403 pKa = 11.84LHH405 pKa = 6.02TMNWWDD411 pKa = 3.85RR412 pKa = 11.84ARR414 pKa = 11.84VLQGINMTLADD425 pKa = 4.12VSPCCRR431 pKa = 11.84ASVSQTSGFDD441 pKa = 3.33PPLQVAAEE449 pKa = 3.95QQLAKK454 pKa = 9.69WAVGVDD460 pKa = 3.54EE461 pKa = 4.82WNPIAVSDD469 pKa = 4.1FLVDD473 pKa = 4.53IPGQLCVYY481 pKa = 10.34
MM1 pKa = 7.84PCMEE5 pKa = 5.44DD6 pKa = 2.8VTMATTVEE14 pKa = 4.24ATDD17 pKa = 3.4SSIWARR23 pKa = 11.84KK24 pKa = 8.33VPHH27 pKa = 6.19SVAIATAEE35 pKa = 4.08QVCLLEE41 pKa = 4.29TVEE44 pKa = 4.32IRR46 pKa = 11.84VPHH49 pKa = 6.7PLHH52 pKa = 6.58PGKK55 pKa = 10.81GLTDD59 pKa = 4.96LNTLAAQFLASKK71 pKa = 9.35PNQWLTQVEE80 pKa = 4.41PTPFQTWVKK89 pKa = 9.52RR90 pKa = 11.84FSPSRR95 pKa = 11.84QTQLEE100 pKa = 3.95AGRR103 pKa = 11.84EE104 pKa = 3.84WVARR108 pKa = 11.84HH109 pKa = 5.68GLLEE113 pKa = 4.65KK114 pKa = 10.7DD115 pKa = 3.44SIMAVFVKK123 pKa = 10.78VEE125 pKa = 4.04TSVTGTDD132 pKa = 3.66PRR134 pKa = 11.84NISPRR139 pKa = 11.84DD140 pKa = 3.41PRR142 pKa = 11.84FLSTLGPFTSRR153 pKa = 11.84LEE155 pKa = 4.02QLAVSCPYY163 pKa = 10.08LVKK166 pKa = 10.88GLDD169 pKa = 3.34TYY171 pKa = 11.6KK172 pKa = 10.57RR173 pKa = 11.84GPLLASYY180 pKa = 8.96WRR182 pKa = 11.84ACVIEE187 pKa = 4.16TDD189 pKa = 4.45FSRR192 pKa = 11.84FDD194 pKa = 3.36MTVSQDD200 pKa = 3.12IINTVEE206 pKa = 3.6RR207 pKa = 11.84SLFRR211 pKa = 11.84RR212 pKa = 11.84CFPEE216 pKa = 4.46GLYY219 pKa = 10.14PDD221 pKa = 5.62LDD223 pKa = 3.98ACLSYY228 pKa = 11.02LDD230 pKa = 4.17TMTGVSDD237 pKa = 3.96LGIMYY242 pKa = 7.69TVSGTRR248 pKa = 11.84ASGDD252 pKa = 3.06AHH254 pKa = 5.77TSIANGYY261 pKa = 8.78INRR264 pKa = 11.84FVIWSCLRR272 pKa = 11.84HH273 pKa = 6.35LDD275 pKa = 3.54PSKK278 pKa = 10.22WDD280 pKa = 3.65SFHH283 pKa = 7.05EE284 pKa = 4.71GDD286 pKa = 5.06DD287 pKa = 3.8GVIFLDD293 pKa = 3.38QDD295 pKa = 3.13IFEE298 pKa = 4.6TARR301 pKa = 11.84EE302 pKa = 4.04CLKK305 pKa = 10.64FASCLGFKK313 pKa = 10.63LKK315 pKa = 10.51VICPEE320 pKa = 4.17SQDD323 pKa = 3.47SAVFCGRR330 pKa = 11.84FTCTTCHH337 pKa = 7.09RR338 pKa = 11.84EE339 pKa = 4.04YY340 pKa = 11.05CDD342 pKa = 4.52LPRR345 pKa = 11.84ALSKK349 pKa = 10.67FHH351 pKa = 6.19TTTKK355 pKa = 10.93NGDD358 pKa = 3.78CKK360 pKa = 11.25SLVLAKK366 pKa = 10.35ALSYY370 pKa = 10.38FATDD374 pKa = 2.36AHH376 pKa = 5.73TPIMGVLCDD385 pKa = 4.56ALIKK389 pKa = 10.62LLSPLVSQRR398 pKa = 11.84LMRR401 pKa = 11.84RR402 pKa = 11.84RR403 pKa = 11.84LHH405 pKa = 6.02TMNWWDD411 pKa = 3.85RR412 pKa = 11.84ARR414 pKa = 11.84VLQGINMTLADD425 pKa = 4.12VSPCCRR431 pKa = 11.84ASVSQTSGFDD441 pKa = 3.33PPLQVAAEE449 pKa = 3.95QQLAKK454 pKa = 9.69WAVGVDD460 pKa = 3.54EE461 pKa = 4.82WNPIAVSDD469 pKa = 4.1FLVDD473 pKa = 4.53IPGQLCVYY481 pKa = 10.34
Molecular weight: 53.77 kDa
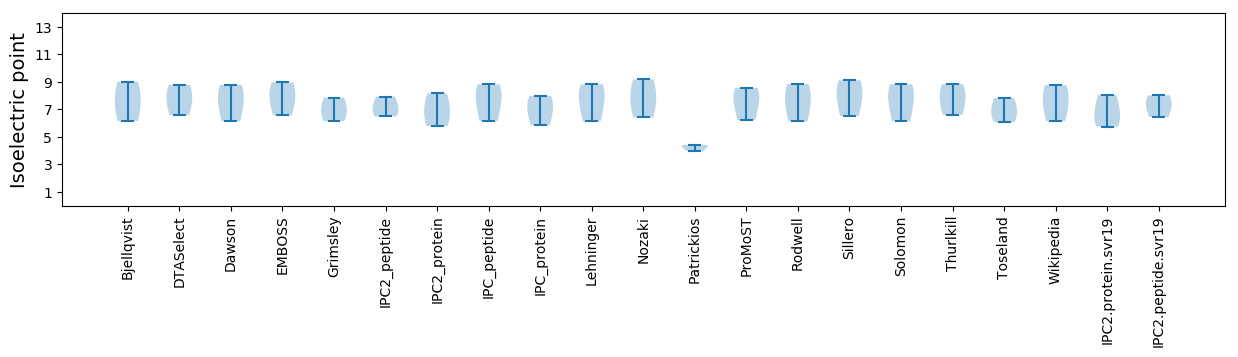
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGS1|A0A1L3KGS1_9VIRU Capsid protein OS=Wenling tombus-like virus 4 OX=1923546 PE=3 SV=1
MM1 pKa = 7.41FFQYY5 pKa = 10.98LLPVLTAIGLLRR17 pKa = 11.84GSTTQLDD24 pKa = 4.13HH25 pKa = 7.07TWFEE29 pKa = 4.32WLGLPLYY36 pKa = 10.56FSLPDD41 pKa = 2.92SWQWVSEE48 pKa = 3.7WCEE51 pKa = 3.72YY52 pKa = 11.05LGLKK56 pKa = 8.75TLMGVPDD63 pKa = 4.22ADD65 pKa = 4.0RR66 pKa = 11.84FQDD69 pKa = 5.43LIDD72 pKa = 4.98DD73 pKa = 4.39CTQLEE78 pKa = 4.48VTISSWKK85 pKa = 8.92LAKK88 pKa = 10.39SLGSFSPVGFSNRR101 pKa = 11.84YY102 pKa = 7.31YY103 pKa = 9.83VCKK106 pKa = 10.54VLIGRR111 pKa = 11.84GYY113 pKa = 9.04VTEE116 pKa = 4.06QSAPLLLRR124 pKa = 11.84LMALPWLVIDD134 pKa = 5.32WILSLGPIALIILTTCTIISVCALALYY161 pKa = 9.83VCYY164 pKa = 10.43ILARR168 pKa = 11.84RR169 pKa = 11.84SNIQVACDD177 pKa = 3.7PPPFTHH183 pKa = 7.54RR184 pKa = 11.84EE185 pKa = 3.99VKK187 pKa = 10.48DD188 pKa = 3.21ILCSYY193 pKa = 8.84FAKK196 pKa = 10.79NKK198 pKa = 10.27LDD200 pKa = 3.69VDD202 pKa = 4.39SVDD205 pKa = 4.33PDD207 pKa = 3.74PTGHH211 pKa = 7.23PKK213 pKa = 10.02LAYY216 pKa = 9.33IRR218 pKa = 11.84RR219 pKa = 11.84QMEE222 pKa = 3.7KK223 pKa = 10.7AMQVFLLRR231 pKa = 11.84LNRR234 pKa = 11.84KK235 pKa = 8.61IRR237 pKa = 11.84DD238 pKa = 3.51VGGSLSRR245 pKa = 11.84SAKK248 pKa = 9.99LLKK251 pKa = 10.27QGSKK255 pKa = 10.38HH256 pKa = 5.3ICFPNLTSADD266 pKa = 3.75KK267 pKa = 10.93EE268 pKa = 4.01RR269 pKa = 11.84LEE271 pKa = 3.97LAKK274 pKa = 10.76RR275 pKa = 11.84NLDD278 pKa = 3.39AEE280 pKa = 4.31AWEE283 pKa = 4.97HH284 pKa = 6.55IGLHH288 pKa = 6.3KK289 pKa = 10.77GDD291 pKa = 4.19EE292 pKa = 4.49CPHH295 pKa = 6.72RR296 pKa = 11.84SLPSYY301 pKa = 7.36MTYY304 pKa = 10.86VDD306 pKa = 4.24FHH308 pKa = 7.06LSPEE312 pKa = 4.19QLCRR316 pKa = 11.84TIKK319 pKa = 10.76SPTLILTHH327 pKa = 7.32DD328 pKa = 3.98FAKK331 pKa = 10.36VDD333 pKa = 3.57KK334 pKa = 11.31DD335 pKa = 3.57GEE337 pKa = 4.11QWFNGEE343 pKa = 4.42CDD345 pKa = 2.96IKK347 pKa = 10.86RR348 pKa = 11.84YY349 pKa = 10.49GDD351 pKa = 4.08FISFQTRR358 pKa = 11.84GGLKK362 pKa = 8.7YY363 pKa = 8.8THH365 pKa = 7.27GYY367 pKa = 8.78HH368 pKa = 6.2AWEE371 pKa = 4.31NEE373 pKa = 3.63GTICTTEE380 pKa = 3.66GTLRR384 pKa = 11.84YY385 pKa = 8.9YY386 pKa = 10.23RR387 pKa = 11.84VYY389 pKa = 9.55EE390 pKa = 3.77AHH392 pKa = 6.82GSIIIYY398 pKa = 9.0ACPVAGEE405 pKa = 3.97YY406 pKa = 9.51TPRR409 pKa = 11.84VDD411 pKa = 4.0NTLCSTAGSADD422 pKa = 3.38AYY424 pKa = 9.94KK425 pKa = 10.85LNNGVEE431 pKa = 4.13AHH433 pKa = 5.95VKK435 pKa = 10.42GDD437 pKa = 3.48VVQVGDD443 pKa = 4.19HH444 pKa = 6.8EE445 pKa = 4.53ISKK448 pKa = 8.97DD449 pKa = 3.08TLYY452 pKa = 10.71RR453 pKa = 11.84VAYY456 pKa = 9.91QMSLLPRR463 pKa = 11.84DD464 pKa = 4.34EE465 pKa = 4.83KK466 pKa = 10.68WLPNCSSVLRR476 pKa = 11.84GRR478 pKa = 11.84FTADD482 pKa = 3.01EE483 pKa = 4.37MPASALHH490 pKa = 7.12DD491 pKa = 3.66SLQFVVMLADD501 pKa = 4.28RR502 pKa = 11.84MAVSYY507 pKa = 8.93RR508 pKa = 11.84QSMIGDD514 pKa = 3.55PQSYY518 pKa = 8.28NFLMRR523 pKa = 11.84RR524 pKa = 11.84VLLLVLNLTSRR535 pKa = 11.84LPDD538 pKa = 3.45TLSAPIVAAFRR549 pKa = 11.84SFRR552 pKa = 11.84RR553 pKa = 11.84FLSGKK558 pKa = 8.0SPRR561 pKa = 11.84HH562 pKa = 5.71ALSSWMWTVHH572 pKa = 6.26RR573 pKa = 11.84VPTYY577 pKa = 10.38EE578 pKa = 4.28VMWDD582 pKa = 3.3NVAAMLIDD590 pKa = 3.91PKK592 pKa = 10.75KK593 pKa = 10.17PNSAKK598 pKa = 10.36QPFPFSGATAVAPTVEE614 pKa = 4.45QPSGDD619 pKa = 3.5TSQDD623 pKa = 2.93EE624 pKa = 4.54GKK626 pKa = 9.42PACRR630 pKa = 11.84DD631 pKa = 3.33RR632 pKa = 11.84KK633 pKa = 9.98EE634 pKa = 4.39SSSSCSPPSPGSSGVGKK651 pKa = 9.89SQAILQPAGLPDD663 pKa = 4.71DD664 pKa = 4.7PANLPATTSANTQALPQGGVGAGKK688 pKa = 10.16KK689 pKa = 9.5KK690 pKa = 10.9AEE692 pKa = 4.45TISRR696 pKa = 11.84TICIHH701 pKa = 5.83AVHH704 pKa = 7.05GGRR707 pKa = 11.84NHH709 pKa = 7.37GDD711 pKa = 3.18DD712 pKa = 3.56CRR714 pKa = 11.84GYY716 pKa = 10.41GLQYY720 pKa = 10.54LGKK723 pKa = 9.41EE724 pKa = 4.39GSALRR729 pKa = 11.84RR730 pKa = 11.84DD731 pKa = 3.98RR732 pKa = 11.84NRR734 pKa = 11.84GASVPIGDD742 pKa = 4.4RR743 pKa = 11.84RR744 pKa = 11.84NSGTPPPPSRR754 pKa = 11.84KK755 pKa = 9.35RR756 pKa = 11.84ADD758 pKa = 3.76RR759 pKa = 11.84SQHH762 pKa = 5.11TGSTISGEE770 pKa = 4.26QTKK773 pKa = 11.21SMVDD777 pKa = 3.15SSGANSLPDD786 pKa = 3.63VGQKK790 pKa = 9.6ILPVKK795 pKa = 9.56TNAARR800 pKa = 11.84GGKK803 pKa = 9.58GVGGATRR810 pKa = 11.84PVGKK814 pKa = 9.98GFHH817 pKa = 6.37NGSVRR822 pKa = 11.84QGGDD826 pKa = 2.88VSHH829 pKa = 7.31RR830 pKa = 11.84NGSSQYY836 pKa = 10.74KK837 pKa = 8.71PARR840 pKa = 11.84SS841 pKa = 3.28
MM1 pKa = 7.41FFQYY5 pKa = 10.98LLPVLTAIGLLRR17 pKa = 11.84GSTTQLDD24 pKa = 4.13HH25 pKa = 7.07TWFEE29 pKa = 4.32WLGLPLYY36 pKa = 10.56FSLPDD41 pKa = 2.92SWQWVSEE48 pKa = 3.7WCEE51 pKa = 3.72YY52 pKa = 11.05LGLKK56 pKa = 8.75TLMGVPDD63 pKa = 4.22ADD65 pKa = 4.0RR66 pKa = 11.84FQDD69 pKa = 5.43LIDD72 pKa = 4.98DD73 pKa = 4.39CTQLEE78 pKa = 4.48VTISSWKK85 pKa = 8.92LAKK88 pKa = 10.39SLGSFSPVGFSNRR101 pKa = 11.84YY102 pKa = 7.31YY103 pKa = 9.83VCKK106 pKa = 10.54VLIGRR111 pKa = 11.84GYY113 pKa = 9.04VTEE116 pKa = 4.06QSAPLLLRR124 pKa = 11.84LMALPWLVIDD134 pKa = 5.32WILSLGPIALIILTTCTIISVCALALYY161 pKa = 9.83VCYY164 pKa = 10.43ILARR168 pKa = 11.84RR169 pKa = 11.84SNIQVACDD177 pKa = 3.7PPPFTHH183 pKa = 7.54RR184 pKa = 11.84EE185 pKa = 3.99VKK187 pKa = 10.48DD188 pKa = 3.21ILCSYY193 pKa = 8.84FAKK196 pKa = 10.79NKK198 pKa = 10.27LDD200 pKa = 3.69VDD202 pKa = 4.39SVDD205 pKa = 4.33PDD207 pKa = 3.74PTGHH211 pKa = 7.23PKK213 pKa = 10.02LAYY216 pKa = 9.33IRR218 pKa = 11.84RR219 pKa = 11.84QMEE222 pKa = 3.7KK223 pKa = 10.7AMQVFLLRR231 pKa = 11.84LNRR234 pKa = 11.84KK235 pKa = 8.61IRR237 pKa = 11.84DD238 pKa = 3.51VGGSLSRR245 pKa = 11.84SAKK248 pKa = 9.99LLKK251 pKa = 10.27QGSKK255 pKa = 10.38HH256 pKa = 5.3ICFPNLTSADD266 pKa = 3.75KK267 pKa = 10.93EE268 pKa = 4.01RR269 pKa = 11.84LEE271 pKa = 3.97LAKK274 pKa = 10.76RR275 pKa = 11.84NLDD278 pKa = 3.39AEE280 pKa = 4.31AWEE283 pKa = 4.97HH284 pKa = 6.55IGLHH288 pKa = 6.3KK289 pKa = 10.77GDD291 pKa = 4.19EE292 pKa = 4.49CPHH295 pKa = 6.72RR296 pKa = 11.84SLPSYY301 pKa = 7.36MTYY304 pKa = 10.86VDD306 pKa = 4.24FHH308 pKa = 7.06LSPEE312 pKa = 4.19QLCRR316 pKa = 11.84TIKK319 pKa = 10.76SPTLILTHH327 pKa = 7.32DD328 pKa = 3.98FAKK331 pKa = 10.36VDD333 pKa = 3.57KK334 pKa = 11.31DD335 pKa = 3.57GEE337 pKa = 4.11QWFNGEE343 pKa = 4.42CDD345 pKa = 2.96IKK347 pKa = 10.86RR348 pKa = 11.84YY349 pKa = 10.49GDD351 pKa = 4.08FISFQTRR358 pKa = 11.84GGLKK362 pKa = 8.7YY363 pKa = 8.8THH365 pKa = 7.27GYY367 pKa = 8.78HH368 pKa = 6.2AWEE371 pKa = 4.31NEE373 pKa = 3.63GTICTTEE380 pKa = 3.66GTLRR384 pKa = 11.84YY385 pKa = 8.9YY386 pKa = 10.23RR387 pKa = 11.84VYY389 pKa = 9.55EE390 pKa = 3.77AHH392 pKa = 6.82GSIIIYY398 pKa = 9.0ACPVAGEE405 pKa = 3.97YY406 pKa = 9.51TPRR409 pKa = 11.84VDD411 pKa = 4.0NTLCSTAGSADD422 pKa = 3.38AYY424 pKa = 9.94KK425 pKa = 10.85LNNGVEE431 pKa = 4.13AHH433 pKa = 5.95VKK435 pKa = 10.42GDD437 pKa = 3.48VVQVGDD443 pKa = 4.19HH444 pKa = 6.8EE445 pKa = 4.53ISKK448 pKa = 8.97DD449 pKa = 3.08TLYY452 pKa = 10.71RR453 pKa = 11.84VAYY456 pKa = 9.91QMSLLPRR463 pKa = 11.84DD464 pKa = 4.34EE465 pKa = 4.83KK466 pKa = 10.68WLPNCSSVLRR476 pKa = 11.84GRR478 pKa = 11.84FTADD482 pKa = 3.01EE483 pKa = 4.37MPASALHH490 pKa = 7.12DD491 pKa = 3.66SLQFVVMLADD501 pKa = 4.28RR502 pKa = 11.84MAVSYY507 pKa = 8.93RR508 pKa = 11.84QSMIGDD514 pKa = 3.55PQSYY518 pKa = 8.28NFLMRR523 pKa = 11.84RR524 pKa = 11.84VLLLVLNLTSRR535 pKa = 11.84LPDD538 pKa = 3.45TLSAPIVAAFRR549 pKa = 11.84SFRR552 pKa = 11.84RR553 pKa = 11.84FLSGKK558 pKa = 8.0SPRR561 pKa = 11.84HH562 pKa = 5.71ALSSWMWTVHH572 pKa = 6.26RR573 pKa = 11.84VPTYY577 pKa = 10.38EE578 pKa = 4.28VMWDD582 pKa = 3.3NVAAMLIDD590 pKa = 3.91PKK592 pKa = 10.75KK593 pKa = 10.17PNSAKK598 pKa = 10.36QPFPFSGATAVAPTVEE614 pKa = 4.45QPSGDD619 pKa = 3.5TSQDD623 pKa = 2.93EE624 pKa = 4.54GKK626 pKa = 9.42PACRR630 pKa = 11.84DD631 pKa = 3.33RR632 pKa = 11.84KK633 pKa = 9.98EE634 pKa = 4.39SSSSCSPPSPGSSGVGKK651 pKa = 9.89SQAILQPAGLPDD663 pKa = 4.71DD664 pKa = 4.7PANLPATTSANTQALPQGGVGAGKK688 pKa = 10.16KK689 pKa = 9.5KK690 pKa = 10.9AEE692 pKa = 4.45TISRR696 pKa = 11.84TICIHH701 pKa = 5.83AVHH704 pKa = 7.05GGRR707 pKa = 11.84NHH709 pKa = 7.37GDD711 pKa = 3.18DD712 pKa = 3.56CRR714 pKa = 11.84GYY716 pKa = 10.41GLQYY720 pKa = 10.54LGKK723 pKa = 9.41EE724 pKa = 4.39GSALRR729 pKa = 11.84RR730 pKa = 11.84DD731 pKa = 3.98RR732 pKa = 11.84NRR734 pKa = 11.84GASVPIGDD742 pKa = 4.4RR743 pKa = 11.84RR744 pKa = 11.84NSGTPPPPSRR754 pKa = 11.84KK755 pKa = 9.35RR756 pKa = 11.84ADD758 pKa = 3.76RR759 pKa = 11.84SQHH762 pKa = 5.11TGSTISGEE770 pKa = 4.26QTKK773 pKa = 11.21SMVDD777 pKa = 3.15SSGANSLPDD786 pKa = 3.63VGQKK790 pKa = 9.6ILPVKK795 pKa = 9.56TNAARR800 pKa = 11.84GGKK803 pKa = 9.58GVGGATRR810 pKa = 11.84PVGKK814 pKa = 9.98GFHH817 pKa = 6.37NGSVRR822 pKa = 11.84QGGDD826 pKa = 2.88VSHH829 pKa = 7.31RR830 pKa = 11.84NGSSQYY836 pKa = 10.74KK837 pKa = 8.71PARR840 pKa = 11.84SS841 pKa = 3.28
Molecular weight: 92.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1793 |
471 |
841 |
597.7 |
66.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.362 ± 0.07 | 2.398 ± 0.653 |
6.525 ± 0.402 | 3.625 ± 0.34 |
3.346 ± 0.45 | 7.418 ± 0.892 |
2.175 ± 0.438 | 4.294 ± 0.069 |
4.573 ± 0.375 | 9.091 ± 0.826 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.175 ± 0.181 | 3.123 ± 0.51 |
5.968 ± 0.214 | 4.239 ± 0.545 |
6.358 ± 0.2 | 8.087 ± 0.631 |
7.25 ± 1.109 | 6.972 ± 0.5 |
1.785 ± 0.213 | 3.235 ± 0.434 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
