
Pseudomonas phage phi6 (Bacteriophage phi-6)
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Vidaverviricetes; Mindivirales; Cystoviridae; Cystovirus
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
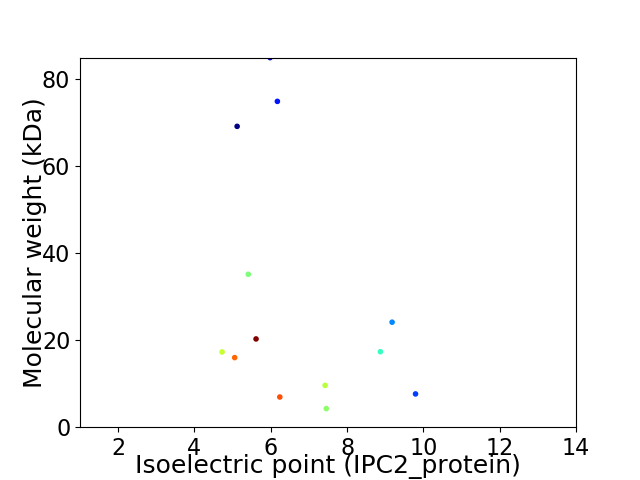
Virtual 2D-PAGE plot for 13 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
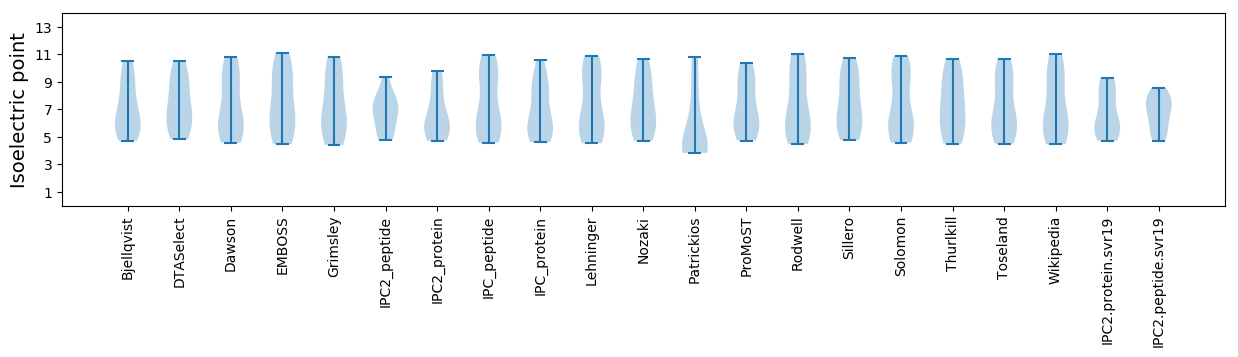
* You can choose from 21 different methods for calculating isoelectric point
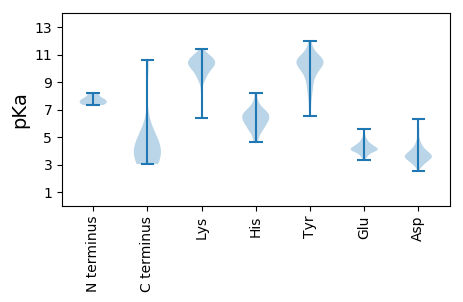
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P11124|RDRP_BPPH6 RNA-directed RNA polymerase OS=Pseudomonas phage phi6 OX=10879 GN=P2 PE=1 SV=3
MM1 pKa = 7.39TLYY4 pKa = 10.67LVPPLDD10 pKa = 4.0SADD13 pKa = 3.84KK14 pKa = 10.13EE15 pKa = 4.4LPALASKK22 pKa = 10.95AGVTLLEE29 pKa = 4.02IEE31 pKa = 4.77FLHH34 pKa = 6.24EE35 pKa = 4.44LWPHH39 pKa = 6.23LSGGQIVIAALNANNLAILNRR60 pKa = 11.84HH61 pKa = 5.44MSTLLVEE68 pKa = 4.65LPVAVMAVPGASYY81 pKa = 10.62RR82 pKa = 11.84SDD84 pKa = 2.83WNMIAHH90 pKa = 7.69ALPSEE95 pKa = 4.06DD96 pKa = 4.97WITLSNKK103 pKa = 7.22MLKK106 pKa = 10.24SGLLANDD113 pKa = 3.88TVQGEE118 pKa = 4.4KK119 pKa = 10.42RR120 pKa = 11.84SGAEE124 pKa = 3.51PLSPNVYY131 pKa = 9.32TDD133 pKa = 3.04ALSRR137 pKa = 11.84LGIATAHH144 pKa = 7.27AIPVEE149 pKa = 4.1PEE151 pKa = 3.57QPFDD155 pKa = 3.81VDD157 pKa = 3.61EE158 pKa = 4.81VSAA161 pKa = 4.47
MM1 pKa = 7.39TLYY4 pKa = 10.67LVPPLDD10 pKa = 4.0SADD13 pKa = 3.84KK14 pKa = 10.13EE15 pKa = 4.4LPALASKK22 pKa = 10.95AGVTLLEE29 pKa = 4.02IEE31 pKa = 4.77FLHH34 pKa = 6.24EE35 pKa = 4.44LWPHH39 pKa = 6.23LSGGQIVIAALNANNLAILNRR60 pKa = 11.84HH61 pKa = 5.44MSTLLVEE68 pKa = 4.65LPVAVMAVPGASYY81 pKa = 10.62RR82 pKa = 11.84SDD84 pKa = 2.83WNMIAHH90 pKa = 7.69ALPSEE95 pKa = 4.06DD96 pKa = 4.97WITLSNKK103 pKa = 7.22MLKK106 pKa = 10.24SGLLANDD113 pKa = 3.88TVQGEE118 pKa = 4.4KK119 pKa = 10.42RR120 pKa = 11.84SGAEE124 pKa = 3.51PLSPNVYY131 pKa = 9.32TDD133 pKa = 3.04ALSRR137 pKa = 11.84LGIATAHH144 pKa = 7.27AIPVEE149 pKa = 4.1PEE151 pKa = 3.57QPFDD155 pKa = 3.81VDD157 pKa = 3.61EE158 pKa = 4.81VSAA161 pKa = 4.47
Molecular weight: 17.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q38563|P14_BPPH6 Protein P14 OS=Pseudomonas phage phi6 OX=10879 GN=P14 PE=4 SV=1
MM1 pKa = 7.33VPLKK5 pKa = 10.56ISTLEE10 pKa = 4.04SQLQPLVKK18 pKa = 10.25LVATEE23 pKa = 4.17TPGALVAYY31 pKa = 10.15ARR33 pKa = 11.84GLSSADD39 pKa = 3.26RR40 pKa = 11.84SRR42 pKa = 11.84LYY44 pKa = 10.62RR45 pKa = 11.84LLRR48 pKa = 11.84SLEE51 pKa = 3.73QAIPKK56 pKa = 9.95LSSAVVSATTLAARR70 pKa = 11.84GLL72 pKa = 3.84
MM1 pKa = 7.33VPLKK5 pKa = 10.56ISTLEE10 pKa = 4.04SQLQPLVKK18 pKa = 10.25LVATEE23 pKa = 4.17TPGALVAYY31 pKa = 10.15ARR33 pKa = 11.84GLSSADD39 pKa = 3.26RR40 pKa = 11.84SRR42 pKa = 11.84LYY44 pKa = 10.62RR45 pKa = 11.84LLRR48 pKa = 11.84SLEE51 pKa = 3.73QAIPKK56 pKa = 9.95LSSAVVSATTLAARR70 pKa = 11.84GLL72 pKa = 3.84
Molecular weight: 7.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3573 |
42 |
769 |
274.8 |
29.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.867 ± 0.696 | 0.756 ± 0.217 |
5.29 ± 0.488 | 4.786 ± 0.634 |
3.554 ± 0.378 | 7.389 ± 0.636 |
1.707 ± 0.261 | 5.01 ± 0.571 |
4.282 ± 0.377 | 9.684 ± 0.57 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.743 ± 0.277 | 3.387 ± 0.401 |
4.898 ± 0.284 | 3.722 ± 0.497 |
5.122 ± 0.639 | 7.305 ± 0.349 |
5.709 ± 0.475 | 8.2 ± 0.702 |
1.539 ± 0.279 | 3.051 ± 0.302 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
