
Chlamydia trachomatis
Taxonomy: cellular organisms; Bacteria; PVC group; Chlamydiae; Chlamydiia; Chlamydiales; Chlamydiaceae;
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
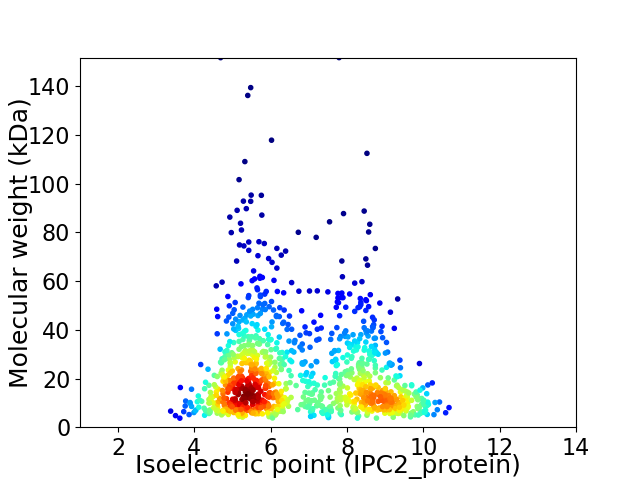
Virtual 2D-PAGE plot for 1102 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A655N281|A0A655N281_CHLTH NIF3-like protein OS=Chlamydia trachomatis OX=813 GN=ERS133252_00387 PE=3 SV=1
MM1 pKa = 7.8VPDD4 pKa = 5.03DD5 pKa = 4.77DD6 pKa = 5.5VGFSSCEE13 pKa = 3.97SPPDD17 pKa = 3.61PEE19 pKa = 5.32LPPKK23 pKa = 10.56LLLFGLTYY31 pKa = 10.45FSFNTDD37 pKa = 2.95PVACTGICSVQDD49 pKa = 3.41CPEE52 pKa = 3.9ASEE55 pKa = 4.15NFLVIGSGVVGVVGVGDD72 pKa = 4.16GVADD76 pKa = 3.7RR77 pKa = 11.84LYY79 pKa = 10.43FLEE82 pKa = 4.35KK83 pKa = 10.72VFVIDD88 pKa = 4.1AVVGPLVAGDD98 pKa = 4.07DD99 pKa = 3.92VASEE103 pKa = 4.45VVFFGEE109 pKa = 4.77DD110 pKa = 3.26GGDD113 pKa = 3.33LGVVSVFGGVTVSSRR128 pKa = 11.84EE129 pKa = 3.83IGSVDD134 pKa = 3.17VGVVLFGWFQVPCMEE149 pKa = 4.59KK150 pKa = 10.22VRR152 pKa = 11.84IPVFF156 pKa = 3.43
MM1 pKa = 7.8VPDD4 pKa = 5.03DD5 pKa = 4.77DD6 pKa = 5.5VGFSSCEE13 pKa = 3.97SPPDD17 pKa = 3.61PEE19 pKa = 5.32LPPKK23 pKa = 10.56LLLFGLTYY31 pKa = 10.45FSFNTDD37 pKa = 2.95PVACTGICSVQDD49 pKa = 3.41CPEE52 pKa = 3.9ASEE55 pKa = 4.15NFLVIGSGVVGVVGVGDD72 pKa = 4.16GVADD76 pKa = 3.7RR77 pKa = 11.84LYY79 pKa = 10.43FLEE82 pKa = 4.35KK83 pKa = 10.72VFVIDD88 pKa = 4.1AVVGPLVAGDD98 pKa = 4.07DD99 pKa = 3.92VASEE103 pKa = 4.45VVFFGEE109 pKa = 4.77DD110 pKa = 3.26GGDD113 pKa = 3.33LGVVSVFGGVTVSSRR128 pKa = 11.84EE129 pKa = 3.83IGSVDD134 pKa = 3.17VGVVLFGWFQVPCMEE149 pKa = 4.59KK150 pKa = 10.22VRR152 pKa = 11.84IPVFF156 pKa = 3.43
Molecular weight: 16.3 kDa
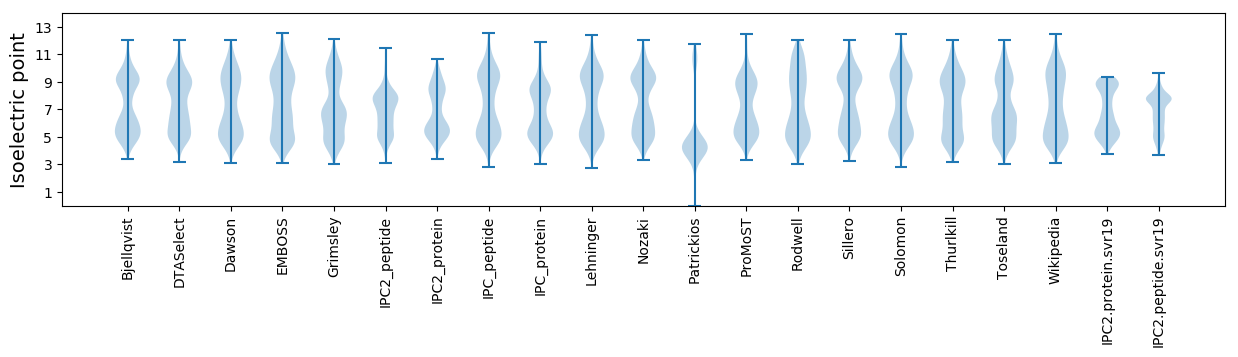
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A655N2I8|A0A655N2I8_CHLTH Uncharacterized protein OS=Chlamydia trachomatis OX=813 GN=ERS133252_00734 PE=4 SV=1
MM1 pKa = 6.76VAVAAAHH8 pKa = 5.32TTTTLAQALTGQGRR22 pKa = 11.84SSSCKK27 pKa = 7.68IWEE30 pKa = 4.96VYY32 pKa = 8.64TQMQDD37 pKa = 3.09FLHH40 pKa = 6.43FRR42 pKa = 11.84THH44 pKa = 4.98FHH46 pKa = 6.32LGFRR50 pKa = 11.84RR51 pKa = 11.84LRR53 pKa = 11.84DD54 pKa = 3.15AKK56 pKa = 8.07TARR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84PHH63 pKa = 5.81SFPWEE68 pKa = 3.96KK69 pKa = 10.82GSALVRR75 pKa = 11.84GGAHH79 pKa = 5.52VNFLVAHH86 pKa = 6.17LNVVRR91 pKa = 11.84HH92 pKa = 6.43ADD94 pKa = 3.48LPLEE98 pKa = 4.12PVEE101 pKa = 4.55RR102 pKa = 11.84HH103 pKa = 5.69DD104 pKa = 4.94QIAAYY109 pKa = 9.73GRR111 pKa = 11.84VIARR115 pKa = 11.84LNEE118 pKa = 3.52IRR120 pKa = 11.84RR121 pKa = 11.84LSVLLLQRR129 pKa = 11.84GAGFLHH135 pKa = 6.74HH136 pKa = 6.69AAEE139 pKa = 4.1RR140 pKa = 11.84HH141 pKa = 5.32EE142 pKa = 4.41PATLQDD148 pKa = 4.22CCQKK152 pKa = 10.8LDD154 pKa = 3.9PLVRR158 pKa = 11.84AAGVCFIDD166 pKa = 4.58PLAGLHH172 pKa = 6.2RR173 pKa = 11.84VLAHH177 pKa = 6.94LGHH180 pKa = 6.06SVRR183 pKa = 11.84TLALNRR189 pKa = 11.84VNLDD193 pKa = 2.7RR194 pKa = 11.84WVVVGAKK201 pKa = 9.78RR202 pKa = 11.84PAPFGHH208 pKa = 6.98PAVMVDD214 pKa = 3.1VVGTSRR220 pKa = 11.84VEE222 pKa = 3.74VALTHH227 pKa = 6.19QHH229 pKa = 6.36DD230 pKa = 4.54CVNVRR235 pKa = 11.84MPLARR240 pKa = 11.84LLVVSDD246 pKa = 4.21FF247 pKa = 3.92
MM1 pKa = 6.76VAVAAAHH8 pKa = 5.32TTTTLAQALTGQGRR22 pKa = 11.84SSSCKK27 pKa = 7.68IWEE30 pKa = 4.96VYY32 pKa = 8.64TQMQDD37 pKa = 3.09FLHH40 pKa = 6.43FRR42 pKa = 11.84THH44 pKa = 4.98FHH46 pKa = 6.32LGFRR50 pKa = 11.84RR51 pKa = 11.84LRR53 pKa = 11.84DD54 pKa = 3.15AKK56 pKa = 8.07TARR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84PHH63 pKa = 5.81SFPWEE68 pKa = 3.96KK69 pKa = 10.82GSALVRR75 pKa = 11.84GGAHH79 pKa = 5.52VNFLVAHH86 pKa = 6.17LNVVRR91 pKa = 11.84HH92 pKa = 6.43ADD94 pKa = 3.48LPLEE98 pKa = 4.12PVEE101 pKa = 4.55RR102 pKa = 11.84HH103 pKa = 5.69DD104 pKa = 4.94QIAAYY109 pKa = 9.73GRR111 pKa = 11.84VIARR115 pKa = 11.84LNEE118 pKa = 3.52IRR120 pKa = 11.84RR121 pKa = 11.84LSVLLLQRR129 pKa = 11.84GAGFLHH135 pKa = 6.74HH136 pKa = 6.69AAEE139 pKa = 4.1RR140 pKa = 11.84HH141 pKa = 5.32EE142 pKa = 4.41PATLQDD148 pKa = 4.22CCQKK152 pKa = 10.8LDD154 pKa = 3.9PLVRR158 pKa = 11.84AAGVCFIDD166 pKa = 4.58PLAGLHH172 pKa = 6.2RR173 pKa = 11.84VLAHH177 pKa = 6.94LGHH180 pKa = 6.06SVRR183 pKa = 11.84TLALNRR189 pKa = 11.84VNLDD193 pKa = 2.7RR194 pKa = 11.84WVVVGAKK201 pKa = 9.78RR202 pKa = 11.84PAPFGHH208 pKa = 6.98PAVMVDD214 pKa = 3.1VVGTSRR220 pKa = 11.84VEE222 pKa = 3.74VALTHH227 pKa = 6.19QHH229 pKa = 6.36DD230 pKa = 4.54CVNVRR235 pKa = 11.84MPLARR240 pKa = 11.84LLVVSDD246 pKa = 4.21FF247 pKa = 3.92
Molecular weight: 27.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
229585 |
31 |
1442 |
208.3 |
23.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.449 ± 0.075 | 1.653 ± 0.039 |
4.554 ± 0.057 | 6.52 ± 0.09 |
4.803 ± 0.068 | 6.267 ± 0.08 |
2.311 ± 0.041 | 6.604 ± 0.071 |
5.878 ± 0.074 | 10.994 ± 0.111 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.35 ± 0.038 | 3.637 ± 0.062 |
4.299 ± 0.066 | 4.129 ± 0.066 |
4.987 ± 0.066 | 7.841 ± 0.087 |
5.065 ± 0.082 | 6.331 ± 0.072 |
1.045 ± 0.03 | 3.254 ± 0.055 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
