
Bovine ephemeral fever virus (BEFV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Ephemerovirus; Bovine fever ephemerovirus
Average proteome isoelectric point is 7.06
Get precalculated fractions of proteins
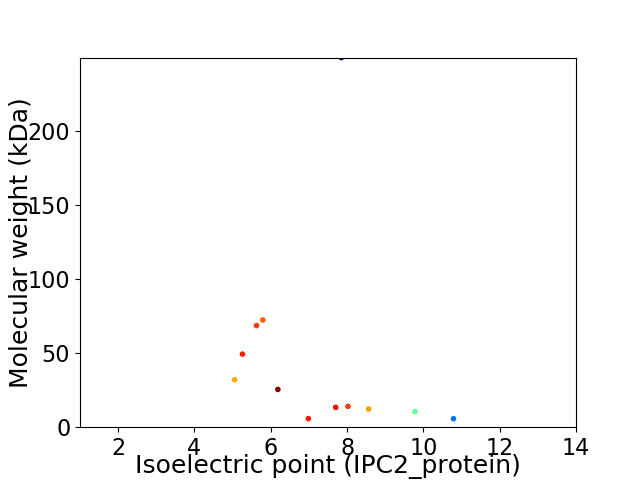
Virtual 2D-PAGE plot for 12 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
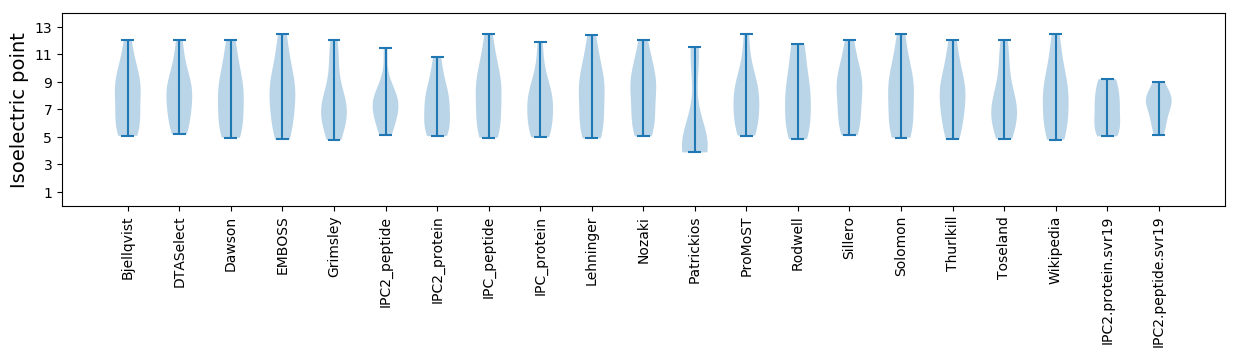
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A0V8W3|A0A0A0V8W3_BEFV GDP polyribonucleotidyltransferase OS=Bovine ephemeral fever virus OX=11303 GN=L PE=4 SV=1
MM1 pKa = 7.31SHH3 pKa = 7.47LKK5 pKa = 9.92PKK7 pKa = 10.41IMTGAYY13 pKa = 9.75DD14 pKa = 3.76AEE16 pKa = 4.35KK17 pKa = 10.59LRR19 pKa = 11.84KK20 pKa = 9.14NLQEE24 pKa = 4.04QIALEE29 pKa = 4.09EE30 pKa = 4.8DD31 pKa = 3.4EE32 pKa = 6.26LEE34 pKa = 4.34NQTDD38 pKa = 3.97FEE40 pKa = 4.85NNKK43 pKa = 9.94EE44 pKa = 3.97EE45 pKa = 4.88SNISNNPLIIKK56 pKa = 8.35QEE58 pKa = 3.87PGIYY62 pKa = 9.52PIEE65 pKa = 4.63INLEE69 pKa = 4.08DD70 pKa = 4.0LEE72 pKa = 4.34KK73 pKa = 11.29ANGIEE78 pKa = 4.02EE79 pKa = 4.12DD80 pKa = 3.85WEE82 pKa = 4.14NSLLKK87 pKa = 10.15IIEE90 pKa = 4.46SSDD93 pKa = 3.47VTPKK97 pKa = 10.86LSWDD101 pKa = 3.87DD102 pKa = 3.58EE103 pKa = 4.76FEE105 pKa = 4.27NDD107 pKa = 3.52TYY109 pKa = 11.17KK110 pKa = 11.06GCNVFTKK117 pKa = 10.25DD118 pKa = 3.82LCNDD122 pKa = 3.38SGNQEE127 pKa = 4.11KK128 pKa = 11.23NNVQIKK134 pKa = 9.72QSSLADD140 pKa = 3.51VAQVLSLFHH149 pKa = 6.48IRR151 pKa = 11.84SDD153 pKa = 3.1VDD155 pKa = 3.55YY156 pKa = 9.08KK157 pKa = 10.23TEE159 pKa = 4.01KK160 pKa = 10.43DD161 pKa = 3.35NKK163 pKa = 9.64GLVRR167 pKa = 11.84IIKK170 pKa = 10.09LSKK173 pKa = 9.22KK174 pKa = 10.33DD175 pKa = 3.8KK176 pKa = 10.76SIEE179 pKa = 4.19SNKK182 pKa = 9.84RR183 pKa = 11.84DD184 pKa = 3.51IVDD187 pKa = 3.55QEE189 pKa = 4.31SDD191 pKa = 2.9KK192 pKa = 10.89HH193 pKa = 5.39TEE195 pKa = 3.99CNSNFDD201 pKa = 3.5AVMDD205 pKa = 3.71QFQKK209 pKa = 11.01GIRR212 pKa = 11.84VKK214 pKa = 10.61KK215 pKa = 10.24RR216 pKa = 11.84FGKK219 pKa = 10.76GYY221 pKa = 10.97VKK223 pKa = 10.44INADD227 pKa = 3.65NMPGTYY233 pKa = 9.53HH234 pKa = 7.52DD235 pKa = 5.06LYY237 pKa = 11.47NVISAVKK244 pKa = 9.83GEE246 pKa = 4.14KK247 pKa = 9.12TVEE250 pKa = 3.53EE251 pKa = 4.31MIRR254 pKa = 11.84YY255 pKa = 8.27LFKK258 pKa = 10.55KK259 pKa = 10.03SKK261 pKa = 9.79SFKK264 pKa = 10.62SISKK268 pKa = 8.5TLNIDD273 pKa = 3.89EE274 pKa = 5.4MILCC278 pKa = 4.91
MM1 pKa = 7.31SHH3 pKa = 7.47LKK5 pKa = 9.92PKK7 pKa = 10.41IMTGAYY13 pKa = 9.75DD14 pKa = 3.76AEE16 pKa = 4.35KK17 pKa = 10.59LRR19 pKa = 11.84KK20 pKa = 9.14NLQEE24 pKa = 4.04QIALEE29 pKa = 4.09EE30 pKa = 4.8DD31 pKa = 3.4EE32 pKa = 6.26LEE34 pKa = 4.34NQTDD38 pKa = 3.97FEE40 pKa = 4.85NNKK43 pKa = 9.94EE44 pKa = 3.97EE45 pKa = 4.88SNISNNPLIIKK56 pKa = 8.35QEE58 pKa = 3.87PGIYY62 pKa = 9.52PIEE65 pKa = 4.63INLEE69 pKa = 4.08DD70 pKa = 4.0LEE72 pKa = 4.34KK73 pKa = 11.29ANGIEE78 pKa = 4.02EE79 pKa = 4.12DD80 pKa = 3.85WEE82 pKa = 4.14NSLLKK87 pKa = 10.15IIEE90 pKa = 4.46SSDD93 pKa = 3.47VTPKK97 pKa = 10.86LSWDD101 pKa = 3.87DD102 pKa = 3.58EE103 pKa = 4.76FEE105 pKa = 4.27NDD107 pKa = 3.52TYY109 pKa = 11.17KK110 pKa = 11.06GCNVFTKK117 pKa = 10.25DD118 pKa = 3.82LCNDD122 pKa = 3.38SGNQEE127 pKa = 4.11KK128 pKa = 11.23NNVQIKK134 pKa = 9.72QSSLADD140 pKa = 3.51VAQVLSLFHH149 pKa = 6.48IRR151 pKa = 11.84SDD153 pKa = 3.1VDD155 pKa = 3.55YY156 pKa = 9.08KK157 pKa = 10.23TEE159 pKa = 4.01KK160 pKa = 10.43DD161 pKa = 3.35NKK163 pKa = 9.64GLVRR167 pKa = 11.84IIKK170 pKa = 10.09LSKK173 pKa = 9.22KK174 pKa = 10.33DD175 pKa = 3.8KK176 pKa = 10.76SIEE179 pKa = 4.19SNKK182 pKa = 9.84RR183 pKa = 11.84DD184 pKa = 3.51IVDD187 pKa = 3.55QEE189 pKa = 4.31SDD191 pKa = 2.9KK192 pKa = 10.89HH193 pKa = 5.39TEE195 pKa = 3.99CNSNFDD201 pKa = 3.5AVMDD205 pKa = 3.71QFQKK209 pKa = 11.01GIRR212 pKa = 11.84VKK214 pKa = 10.61KK215 pKa = 10.24RR216 pKa = 11.84FGKK219 pKa = 10.76GYY221 pKa = 10.97VKK223 pKa = 10.44INADD227 pKa = 3.65NMPGTYY233 pKa = 9.53HH234 pKa = 7.52DD235 pKa = 5.06LYY237 pKa = 11.47NVISAVKK244 pKa = 9.83GEE246 pKa = 4.14KK247 pKa = 9.12TVEE250 pKa = 3.53EE251 pKa = 4.31MIRR254 pKa = 11.84YY255 pKa = 8.27LFKK258 pKa = 10.55KK259 pKa = 10.03SKK261 pKa = 9.79SFKK264 pKa = 10.62SISKK268 pKa = 8.5TLNIDD273 pKa = 3.89EE274 pKa = 5.4MILCC278 pKa = 4.91
Molecular weight: 32.0 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A0VBA2|A0A0A0VBA2_BEFV Isoform of A0A0A0V4V7 Alpha 3 protein OS=Bovine ephemeral fever virus OX=11303 GN=alpha PE=4 SV=1
MM1 pKa = 7.65EE2 pKa = 6.12KK3 pKa = 10.88GLLSNFWNDD12 pKa = 2.85FKK14 pKa = 11.19QWSEE18 pKa = 3.81DD19 pKa = 3.21RR20 pKa = 11.84KK21 pKa = 10.5VEE23 pKa = 4.54IVNWWSNLEE32 pKa = 4.17SKK34 pKa = 10.51VRR36 pKa = 11.84LGFWIILIILLGILAIRR53 pKa = 11.84IAIKK57 pKa = 10.34VYY59 pKa = 10.12QCVKK63 pKa = 9.78LTTKK67 pKa = 10.14GVKK70 pKa = 9.62KK71 pKa = 10.09IKK73 pKa = 10.25RR74 pKa = 11.84VIKK77 pKa = 10.08KK78 pKa = 9.4KK79 pKa = 10.62RR80 pKa = 11.84SIKK83 pKa = 10.22KK84 pKa = 9.1YY85 pKa = 10.38RR86 pKa = 11.84KK87 pKa = 6.76TT88 pKa = 3.33
MM1 pKa = 7.65EE2 pKa = 6.12KK3 pKa = 10.88GLLSNFWNDD12 pKa = 2.85FKK14 pKa = 11.19QWSEE18 pKa = 3.81DD19 pKa = 3.21RR20 pKa = 11.84KK21 pKa = 10.5VEE23 pKa = 4.54IVNWWSNLEE32 pKa = 4.17SKK34 pKa = 10.51VRR36 pKa = 11.84LGFWIILIILLGILAIRR53 pKa = 11.84IAIKK57 pKa = 10.34VYY59 pKa = 10.12QCVKK63 pKa = 9.78LTTKK67 pKa = 10.14GVKK70 pKa = 9.62KK71 pKa = 10.09IKK73 pKa = 10.25RR74 pKa = 11.84VIKK77 pKa = 10.08KK78 pKa = 9.4KK79 pKa = 10.62RR80 pKa = 11.84SIKK83 pKa = 10.22KK84 pKa = 9.1YY85 pKa = 10.38RR86 pKa = 11.84KK87 pKa = 6.76TT88 pKa = 3.33
Molecular weight: 10.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4809 |
48 |
2144 |
400.8 |
46.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
2.807 ± 0.482 | 1.975 ± 0.22 |
5.614 ± 0.369 | 7.798 ± 0.419 |
4.034 ± 0.203 | 5.511 ± 0.312 |
2.371 ± 0.227 | 8.526 ± 0.435 |
8.692 ± 0.519 | 10.002 ± 0.79 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.599 ± 0.343 | 6.758 ± 0.264 |
3.098 ± 0.224 | 2.89 ± 0.15 |
5.053 ± 0.324 | 7.216 ± 0.515 |
4.408 ± 0.328 | 4.658 ± 0.396 |
1.871 ± 0.209 | 4.055 ± 0.337 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
