
Actinoplanes atraurantiacus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micromonosporales; Micromonosporaceae; Actinoplanes
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
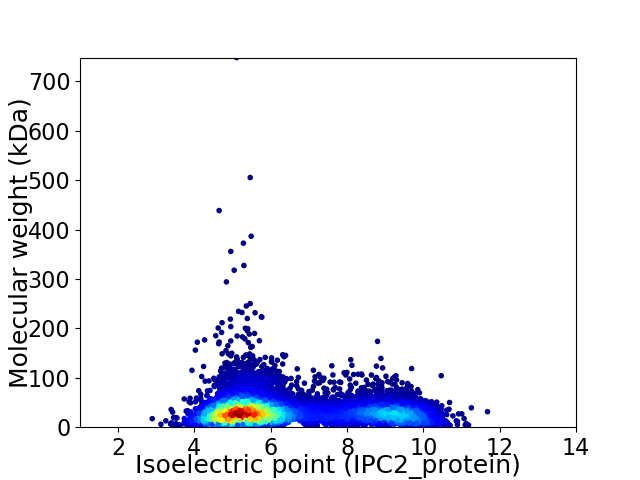
Virtual 2D-PAGE plot for 10240 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A285ICT6|A0A285ICT6_9ACTN Histidine triad (HIT) family protein OS=Actinoplanes atraurantiacus OX=1036182 GN=SAMN05421748_107257 PE=4 SV=1
MM1 pKa = 7.68TDD3 pKa = 2.63HH4 pKa = 7.43HH5 pKa = 7.39DD6 pKa = 4.22FGDD9 pKa = 3.94HH10 pKa = 6.82EE11 pKa = 5.46DD12 pKa = 3.99DD13 pKa = 5.04FGVFDD18 pKa = 3.97DD19 pKa = 4.5HH20 pKa = 7.04QLPEE24 pKa = 4.27HH25 pKa = 6.86AEE27 pKa = 3.8PLPFEE32 pKa = 4.39DD33 pKa = 3.31HH34 pKa = 7.09FEE36 pKa = 4.43YY37 pKa = 10.8EE38 pKa = 4.29VDD40 pKa = 3.49DD41 pKa = 3.55HH42 pKa = 6.46HH43 pKa = 7.3TYY45 pKa = 10.9DD46 pKa = 4.01EE47 pKa = 4.77PVHH50 pKa = 6.44EE51 pKa = 4.63PAEE54 pKa = 4.35HH55 pKa = 6.45VFDD58 pKa = 4.29EE59 pKa = 5.0PEE61 pKa = 4.7HH62 pKa = 6.11EE63 pKa = 4.56VAEE66 pKa = 4.65LPADD70 pKa = 3.96DD71 pKa = 5.01PVDD74 pKa = 3.7VFPPPVHH81 pKa = 7.36VGEE84 pKa = 4.61LPEE87 pKa = 4.54PVDD90 pKa = 3.63GFPWIDD96 pKa = 3.28TGSLGVVPADD106 pKa = 3.41ALVTPVDD113 pKa = 3.51PVQPQEE119 pKa = 3.9LADD122 pKa = 3.8YY123 pKa = 10.57AGEE126 pKa = 3.92QLPPGVDD133 pKa = 3.14PWAALADD140 pKa = 4.08SEE142 pKa = 5.58DD143 pKa = 4.11PATSTLARR151 pKa = 11.84WWQEE155 pKa = 3.5NN156 pKa = 3.32
MM1 pKa = 7.68TDD3 pKa = 2.63HH4 pKa = 7.43HH5 pKa = 7.39DD6 pKa = 4.22FGDD9 pKa = 3.94HH10 pKa = 6.82EE11 pKa = 5.46DD12 pKa = 3.99DD13 pKa = 5.04FGVFDD18 pKa = 3.97DD19 pKa = 4.5HH20 pKa = 7.04QLPEE24 pKa = 4.27HH25 pKa = 6.86AEE27 pKa = 3.8PLPFEE32 pKa = 4.39DD33 pKa = 3.31HH34 pKa = 7.09FEE36 pKa = 4.43YY37 pKa = 10.8EE38 pKa = 4.29VDD40 pKa = 3.49DD41 pKa = 3.55HH42 pKa = 6.46HH43 pKa = 7.3TYY45 pKa = 10.9DD46 pKa = 4.01EE47 pKa = 4.77PVHH50 pKa = 6.44EE51 pKa = 4.63PAEE54 pKa = 4.35HH55 pKa = 6.45VFDD58 pKa = 4.29EE59 pKa = 5.0PEE61 pKa = 4.7HH62 pKa = 6.11EE63 pKa = 4.56VAEE66 pKa = 4.65LPADD70 pKa = 3.96DD71 pKa = 5.01PVDD74 pKa = 3.7VFPPPVHH81 pKa = 7.36VGEE84 pKa = 4.61LPEE87 pKa = 4.54PVDD90 pKa = 3.63GFPWIDD96 pKa = 3.28TGSLGVVPADD106 pKa = 3.41ALVTPVDD113 pKa = 3.51PVQPQEE119 pKa = 3.9LADD122 pKa = 3.8YY123 pKa = 10.57AGEE126 pKa = 3.92QLPPGVDD133 pKa = 3.14PWAALADD140 pKa = 4.08SEE142 pKa = 5.58DD143 pKa = 4.11PATSTLARR151 pKa = 11.84WWQEE155 pKa = 3.5NN156 pKa = 3.32
Molecular weight: 17.43 kDa
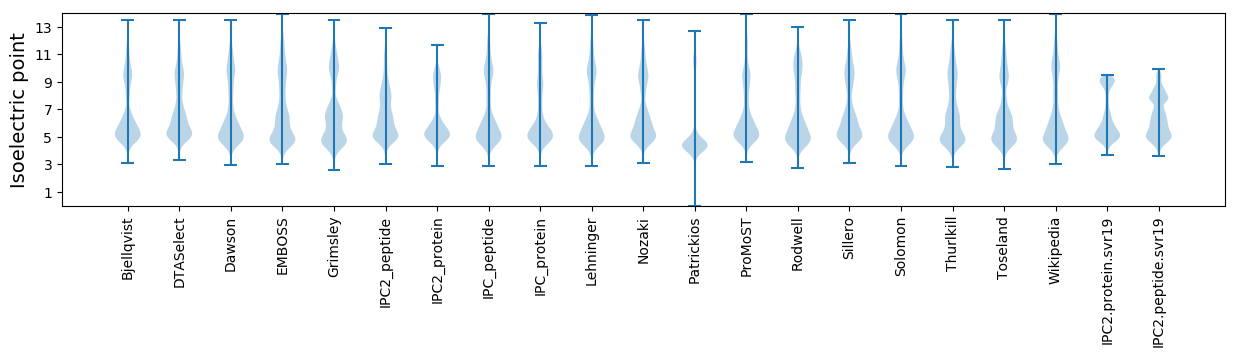
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A285FV58|A0A285FV58_9ACTN Uncharacterized protein OS=Actinoplanes atraurantiacus OX=1036182 GN=SAMN05421748_1011287 PE=4 SV=1
MM1 pKa = 7.05VPSRR5 pKa = 11.84QVPLRR10 pKa = 11.84RR11 pKa = 11.84LRR13 pKa = 11.84PRR15 pKa = 11.84PRR17 pKa = 11.84PPPAVLRR24 pKa = 11.84PARR27 pKa = 11.84RR28 pKa = 11.84HH29 pKa = 4.71RR30 pKa = 11.84LARR33 pKa = 11.84LRR35 pKa = 11.84PPRR38 pKa = 11.84VPSRR42 pKa = 11.84RR43 pKa = 11.84AGPAPPPVALPAPRR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84PPRR63 pKa = 11.84RR64 pKa = 11.84PALSRR69 pKa = 11.84VPSRR73 pKa = 11.84RR74 pKa = 11.84FPAVRR79 pKa = 11.84APRR82 pKa = 11.84PARR85 pKa = 11.84SPSRR89 pKa = 11.84PAPTTSRR96 pKa = 11.84SRR98 pKa = 11.84RR99 pKa = 11.84PRR101 pKa = 11.84PVPRR105 pKa = 11.84RR106 pKa = 11.84SRR108 pKa = 11.84PSRR111 pKa = 11.84RR112 pKa = 11.84PRR114 pKa = 11.84SRR116 pKa = 11.84PLRR119 pKa = 11.84PPARR123 pKa = 11.84VTPSAGRR130 pKa = 11.84SSPRR134 pKa = 11.84PTAARR139 pKa = 11.84PVAPDD144 pKa = 3.92RR145 pKa = 11.84VPAPARR151 pKa = 11.84FRR153 pKa = 11.84RR154 pKa = 11.84ARR156 pKa = 11.84VRR158 pKa = 11.84PRR160 pKa = 11.84PAPPRR165 pKa = 11.84RR166 pKa = 11.84VAAVLRR172 pKa = 11.84PARR175 pKa = 11.84VPAPPVATPRR185 pKa = 11.84ARR187 pKa = 11.84RR188 pKa = 11.84RR189 pKa = 11.84VARR192 pKa = 11.84VTTRR196 pKa = 11.84SASRR200 pKa = 11.84VATPPVPRR208 pKa = 11.84RR209 pKa = 11.84LPCPGPTSRR218 pKa = 11.84ACPRR222 pKa = 11.84GRR224 pKa = 11.84ARR226 pKa = 11.84PRR228 pKa = 11.84CPRR231 pKa = 11.84GPAPRR236 pKa = 11.84PCPRR240 pKa = 11.84SVRR243 pKa = 11.84PAPAVRR249 pKa = 11.84VAAPVAVAAPVAAAVVAVPVAASVPVAAAVPVAASVPVAAAVPVVAVATAVVPAAAVAVATAAVPVVAAVLRR321 pKa = 11.84PVPALRR327 pKa = 11.84VVPVAAVAVVAAVPRR342 pKa = 11.84VPSGVRR348 pKa = 11.84AAGRR352 pKa = 11.84PAVASRR358 pKa = 11.84RR359 pKa = 11.84SSGVKK364 pKa = 10.17SSTTCPPRR372 pKa = 11.84RR373 pKa = 3.6
MM1 pKa = 7.05VPSRR5 pKa = 11.84QVPLRR10 pKa = 11.84RR11 pKa = 11.84LRR13 pKa = 11.84PRR15 pKa = 11.84PRR17 pKa = 11.84PPPAVLRR24 pKa = 11.84PARR27 pKa = 11.84RR28 pKa = 11.84HH29 pKa = 4.71RR30 pKa = 11.84LARR33 pKa = 11.84LRR35 pKa = 11.84PPRR38 pKa = 11.84VPSRR42 pKa = 11.84RR43 pKa = 11.84AGPAPPPVALPAPRR57 pKa = 11.84RR58 pKa = 11.84RR59 pKa = 11.84RR60 pKa = 11.84PPRR63 pKa = 11.84RR64 pKa = 11.84PALSRR69 pKa = 11.84VPSRR73 pKa = 11.84RR74 pKa = 11.84FPAVRR79 pKa = 11.84APRR82 pKa = 11.84PARR85 pKa = 11.84SPSRR89 pKa = 11.84PAPTTSRR96 pKa = 11.84SRR98 pKa = 11.84RR99 pKa = 11.84PRR101 pKa = 11.84PVPRR105 pKa = 11.84RR106 pKa = 11.84SRR108 pKa = 11.84PSRR111 pKa = 11.84RR112 pKa = 11.84PRR114 pKa = 11.84SRR116 pKa = 11.84PLRR119 pKa = 11.84PPARR123 pKa = 11.84VTPSAGRR130 pKa = 11.84SSPRR134 pKa = 11.84PTAARR139 pKa = 11.84PVAPDD144 pKa = 3.92RR145 pKa = 11.84VPAPARR151 pKa = 11.84FRR153 pKa = 11.84RR154 pKa = 11.84ARR156 pKa = 11.84VRR158 pKa = 11.84PRR160 pKa = 11.84PAPPRR165 pKa = 11.84RR166 pKa = 11.84VAAVLRR172 pKa = 11.84PARR175 pKa = 11.84VPAPPVATPRR185 pKa = 11.84ARR187 pKa = 11.84RR188 pKa = 11.84RR189 pKa = 11.84VARR192 pKa = 11.84VTTRR196 pKa = 11.84SASRR200 pKa = 11.84VATPPVPRR208 pKa = 11.84RR209 pKa = 11.84LPCPGPTSRR218 pKa = 11.84ACPRR222 pKa = 11.84GRR224 pKa = 11.84ARR226 pKa = 11.84PRR228 pKa = 11.84CPRR231 pKa = 11.84GPAPRR236 pKa = 11.84PCPRR240 pKa = 11.84SVRR243 pKa = 11.84PAPAVRR249 pKa = 11.84VAAPVAVAAPVAAAVVAVPVAASVPVAAAVPVAASVPVAAAVPVVAVATAVVPAAAVAVATAAVPVVAAVLRR321 pKa = 11.84PVPALRR327 pKa = 11.84VVPVAAVAVVAAVPRR342 pKa = 11.84VPSGVRR348 pKa = 11.84AAGRR352 pKa = 11.84PAVASRR358 pKa = 11.84RR359 pKa = 11.84SSGVKK364 pKa = 10.17SSTTCPPRR372 pKa = 11.84RR373 pKa = 3.6
Molecular weight: 39.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3323307 |
24 |
7153 |
324.5 |
34.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.908 ± 0.039 | 0.665 ± 0.006 |
5.969 ± 0.021 | 5.221 ± 0.025 |
2.834 ± 0.015 | 9.2 ± 0.025 |
2.064 ± 0.012 | 3.664 ± 0.017 |
2.066 ± 0.018 | 10.334 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.734 ± 0.009 | 1.968 ± 0.016 |
6.06 ± 0.022 | 2.771 ± 0.014 |
7.857 ± 0.03 | 5.27 ± 0.022 |
6.18 ± 0.027 | 8.522 ± 0.023 |
1.593 ± 0.01 | 2.121 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
