
Hubei sobemo-like virus 7
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.42
Get precalculated fractions of proteins
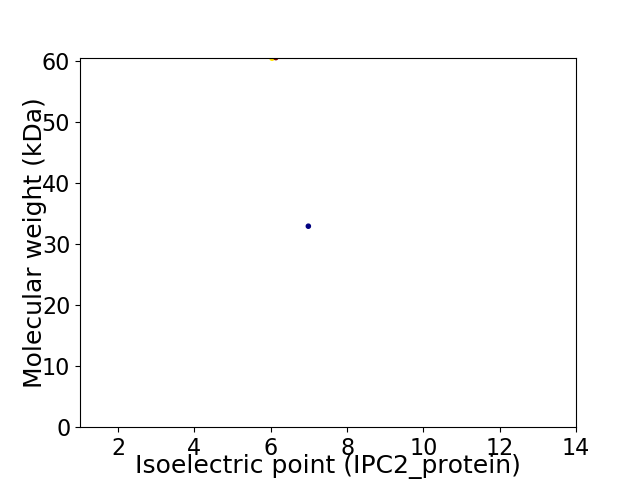
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KEN9|A0A1L3KEN9_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 7 OX=1923240 PE=4 SV=1
MM1 pKa = 7.1FVSSNVKK8 pKa = 10.26AMEE11 pKa = 4.23QIEE14 pKa = 4.51MVLTMMMVSLVVTLSVKK31 pKa = 9.82MLTEE35 pKa = 3.84LVRR38 pKa = 11.84WYY40 pKa = 10.67LRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84AQYY47 pKa = 10.93LYY49 pKa = 11.03AKK51 pKa = 9.54KK52 pKa = 10.34QEE54 pKa = 4.27DD55 pKa = 3.84LGRR58 pKa = 11.84LPEE61 pKa = 4.11SMRR64 pKa = 11.84EE65 pKa = 3.95GSDD68 pKa = 3.37LVTVPLANFPFVVGLYY84 pKa = 8.78SVRR87 pKa = 11.84RR88 pKa = 11.84QGAAEE93 pKa = 3.95KK94 pKa = 10.43VYY96 pKa = 10.88LGTGTLLEE104 pKa = 4.58GWLVTAGHH112 pKa = 5.38VVEE115 pKa = 4.89TAAGRR120 pKa = 11.84DD121 pKa = 3.92MFALTYY127 pKa = 10.67DD128 pKa = 3.9GVYY131 pKa = 10.45HH132 pKa = 6.93EE133 pKa = 5.86LGQFHH138 pKa = 7.32TIYY141 pKa = 10.27TDD143 pKa = 3.46LVMTKK148 pKa = 10.25APQGYY153 pKa = 9.28KK154 pKa = 9.76SGRR157 pKa = 11.84VEE159 pKa = 4.02ALQLASYY166 pKa = 8.99AQLVAARR173 pKa = 11.84GTANSSMGILKK184 pKa = 10.05HH185 pKa = 5.67DD186 pKa = 3.68TQVAFGFCSYY196 pKa = 10.52SGSSVQGFSGAPYY209 pKa = 8.9TNGSKK214 pKa = 10.43VLAIHH219 pKa = 6.92LGGGTSGNYY228 pKa = 9.69GYY230 pKa = 10.46SASFVMAQIHH240 pKa = 5.46VYY242 pKa = 9.77KK243 pKa = 10.39RR244 pKa = 11.84PEE246 pKa = 3.97SSEE249 pKa = 4.04LEE251 pKa = 4.01AMEE254 pKa = 5.0RR255 pKa = 11.84AMKK258 pKa = 10.03SAKK261 pKa = 10.26KK262 pKa = 10.24EE263 pKa = 4.14DD264 pKa = 3.82IEE266 pKa = 4.23WEE268 pKa = 4.0RR269 pKa = 11.84GLDD272 pKa = 3.43EE273 pKa = 3.89TRR275 pKa = 11.84VRR277 pKa = 11.84VGGRR281 pKa = 11.84YY282 pKa = 7.37FTLDD286 pKa = 2.97NDD288 pKa = 4.61EE289 pKa = 4.03FDD291 pKa = 3.82EE292 pKa = 4.35WSEE295 pKa = 3.92LNEE298 pKa = 4.1FGEE301 pKa = 4.18WFYY304 pKa = 11.68DD305 pKa = 3.64EE306 pKa = 5.06EE307 pKa = 5.28PEE309 pKa = 4.46SKK311 pKa = 10.24RR312 pKa = 11.84KK313 pKa = 9.44RR314 pKa = 11.84FRR316 pKa = 11.84VKK318 pKa = 10.27RR319 pKa = 11.84KK320 pKa = 9.4AVDD323 pKa = 3.56YY324 pKa = 10.35EE325 pKa = 4.16EE326 pKa = 5.38PDD328 pKa = 3.61YY329 pKa = 11.3EE330 pKa = 4.33PEE332 pKa = 4.2GVEE335 pKa = 5.57DD336 pKa = 4.83PFLEE340 pKa = 4.44SKK342 pKa = 10.21PPKK345 pKa = 9.89DD346 pKa = 3.67SSSGALEE353 pKa = 3.86KK354 pKa = 10.51RR355 pKa = 11.84IEE357 pKa = 3.98LLAEE361 pKa = 4.53NIASIQSLLTSQSQDD376 pKa = 2.83MGGLKK381 pKa = 9.96EE382 pKa = 4.51RR383 pKa = 11.84LNKK386 pKa = 10.14LDD388 pKa = 3.72TACVSMRR395 pKa = 11.84NIEE398 pKa = 4.39SKK400 pKa = 10.73LSEE403 pKa = 4.27TFASQLTMSVNTLEE417 pKa = 4.37QRR419 pKa = 11.84LTAQIKK425 pKa = 9.52SVSDD429 pKa = 3.52ARR431 pKa = 11.84PHH433 pKa = 6.22PEE435 pKa = 3.64KK436 pKa = 10.02TASTNGSDD444 pKa = 3.36TSSPPSIPEE453 pKa = 3.82VPQVSVSSGSTQPSVMRR470 pKa = 11.84WATMDD475 pKa = 3.41SDD477 pKa = 4.09FATMLEE483 pKa = 3.87WRR485 pKa = 11.84NSVDD489 pKa = 3.19RR490 pKa = 11.84SSPSYY495 pKa = 8.07PHH497 pKa = 6.18WRR499 pKa = 11.84EE500 pKa = 3.29EE501 pKa = 3.98YY502 pKa = 7.48MTQKK506 pKa = 10.68GFSTEE511 pKa = 3.77QKK513 pKa = 8.86KK514 pKa = 10.81ALVVRR519 pKa = 11.84LKK521 pKa = 10.45NYY523 pKa = 9.69KK524 pKa = 9.78VSSQRR529 pKa = 11.84KK530 pKa = 4.8VARR533 pKa = 11.84KK534 pKa = 9.35AKK536 pKa = 10.39SAIQQ540 pKa = 3.27
MM1 pKa = 7.1FVSSNVKK8 pKa = 10.26AMEE11 pKa = 4.23QIEE14 pKa = 4.51MVLTMMMVSLVVTLSVKK31 pKa = 9.82MLTEE35 pKa = 3.84LVRR38 pKa = 11.84WYY40 pKa = 10.67LRR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84AQYY47 pKa = 10.93LYY49 pKa = 11.03AKK51 pKa = 9.54KK52 pKa = 10.34QEE54 pKa = 4.27DD55 pKa = 3.84LGRR58 pKa = 11.84LPEE61 pKa = 4.11SMRR64 pKa = 11.84EE65 pKa = 3.95GSDD68 pKa = 3.37LVTVPLANFPFVVGLYY84 pKa = 8.78SVRR87 pKa = 11.84RR88 pKa = 11.84QGAAEE93 pKa = 3.95KK94 pKa = 10.43VYY96 pKa = 10.88LGTGTLLEE104 pKa = 4.58GWLVTAGHH112 pKa = 5.38VVEE115 pKa = 4.89TAAGRR120 pKa = 11.84DD121 pKa = 3.92MFALTYY127 pKa = 10.67DD128 pKa = 3.9GVYY131 pKa = 10.45HH132 pKa = 6.93EE133 pKa = 5.86LGQFHH138 pKa = 7.32TIYY141 pKa = 10.27TDD143 pKa = 3.46LVMTKK148 pKa = 10.25APQGYY153 pKa = 9.28KK154 pKa = 9.76SGRR157 pKa = 11.84VEE159 pKa = 4.02ALQLASYY166 pKa = 8.99AQLVAARR173 pKa = 11.84GTANSSMGILKK184 pKa = 10.05HH185 pKa = 5.67DD186 pKa = 3.68TQVAFGFCSYY196 pKa = 10.52SGSSVQGFSGAPYY209 pKa = 8.9TNGSKK214 pKa = 10.43VLAIHH219 pKa = 6.92LGGGTSGNYY228 pKa = 9.69GYY230 pKa = 10.46SASFVMAQIHH240 pKa = 5.46VYY242 pKa = 9.77KK243 pKa = 10.39RR244 pKa = 11.84PEE246 pKa = 3.97SSEE249 pKa = 4.04LEE251 pKa = 4.01AMEE254 pKa = 5.0RR255 pKa = 11.84AMKK258 pKa = 10.03SAKK261 pKa = 10.26KK262 pKa = 10.24EE263 pKa = 4.14DD264 pKa = 3.82IEE266 pKa = 4.23WEE268 pKa = 4.0RR269 pKa = 11.84GLDD272 pKa = 3.43EE273 pKa = 3.89TRR275 pKa = 11.84VRR277 pKa = 11.84VGGRR281 pKa = 11.84YY282 pKa = 7.37FTLDD286 pKa = 2.97NDD288 pKa = 4.61EE289 pKa = 4.03FDD291 pKa = 3.82EE292 pKa = 4.35WSEE295 pKa = 3.92LNEE298 pKa = 4.1FGEE301 pKa = 4.18WFYY304 pKa = 11.68DD305 pKa = 3.64EE306 pKa = 5.06EE307 pKa = 5.28PEE309 pKa = 4.46SKK311 pKa = 10.24RR312 pKa = 11.84KK313 pKa = 9.44RR314 pKa = 11.84FRR316 pKa = 11.84VKK318 pKa = 10.27RR319 pKa = 11.84KK320 pKa = 9.4AVDD323 pKa = 3.56YY324 pKa = 10.35EE325 pKa = 4.16EE326 pKa = 5.38PDD328 pKa = 3.61YY329 pKa = 11.3EE330 pKa = 4.33PEE332 pKa = 4.2GVEE335 pKa = 5.57DD336 pKa = 4.83PFLEE340 pKa = 4.44SKK342 pKa = 10.21PPKK345 pKa = 9.89DD346 pKa = 3.67SSSGALEE353 pKa = 3.86KK354 pKa = 10.51RR355 pKa = 11.84IEE357 pKa = 3.98LLAEE361 pKa = 4.53NIASIQSLLTSQSQDD376 pKa = 2.83MGGLKK381 pKa = 9.96EE382 pKa = 4.51RR383 pKa = 11.84LNKK386 pKa = 10.14LDD388 pKa = 3.72TACVSMRR395 pKa = 11.84NIEE398 pKa = 4.39SKK400 pKa = 10.73LSEE403 pKa = 4.27TFASQLTMSVNTLEE417 pKa = 4.37QRR419 pKa = 11.84LTAQIKK425 pKa = 9.52SVSDD429 pKa = 3.52ARR431 pKa = 11.84PHH433 pKa = 6.22PEE435 pKa = 3.64KK436 pKa = 10.02TASTNGSDD444 pKa = 3.36TSSPPSIPEE453 pKa = 3.82VPQVSVSSGSTQPSVMRR470 pKa = 11.84WATMDD475 pKa = 3.41SDD477 pKa = 4.09FATMLEE483 pKa = 3.87WRR485 pKa = 11.84NSVDD489 pKa = 3.19RR490 pKa = 11.84SSPSYY495 pKa = 8.07PHH497 pKa = 6.18WRR499 pKa = 11.84EE500 pKa = 3.29EE501 pKa = 3.98YY502 pKa = 7.48MTQKK506 pKa = 10.68GFSTEE511 pKa = 3.77QKK513 pKa = 8.86KK514 pKa = 10.81ALVVRR519 pKa = 11.84LKK521 pKa = 10.45NYY523 pKa = 9.69KK524 pKa = 9.78VSSQRR529 pKa = 11.84KK530 pKa = 4.8VARR533 pKa = 11.84KK534 pKa = 9.35AKK536 pKa = 10.39SAIQQ540 pKa = 3.27
Molecular weight: 60.46 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KEN9|A0A1L3KEN9_9VIRU Uncharacterized protein OS=Hubei sobemo-like virus 7 OX=1923240 PE=4 SV=1
MM1 pKa = 7.77VDD3 pKa = 3.23RR4 pKa = 11.84CLMHH8 pKa = 6.86PFIQSEE14 pKa = 4.03IRR16 pKa = 11.84NPMDD20 pKa = 3.43VVSKK24 pKa = 10.94AGWAPLPAGYY34 pKa = 10.28QRR36 pKa = 11.84MIAEE40 pKa = 4.38FPEE43 pKa = 4.18KK44 pKa = 9.67QAYY47 pKa = 10.11AIDD50 pKa = 3.75KK51 pKa = 10.41KK52 pKa = 10.9LWDD55 pKa = 3.4WTMPGWVVLEE65 pKa = 3.92YY66 pKa = 11.14VLAKK70 pKa = 10.35LRR72 pKa = 11.84QCRR75 pKa = 11.84EE76 pKa = 3.71HH77 pKa = 6.85SLLWEE82 pKa = 3.71WMVWRR87 pKa = 11.84RR88 pKa = 11.84IYY90 pKa = 11.16YY91 pKa = 9.86SFGPGAVLAEE101 pKa = 4.0PTGAIWRR108 pKa = 11.84QTFWGLMKK116 pKa = 10.57SGSLLTLSMNGASQYY131 pKa = 10.93LEE133 pKa = 4.8HH134 pKa = 6.87ALAWLRR140 pKa = 11.84MLDD143 pKa = 3.98GGPISWPPRR152 pKa = 11.84MWAMGDD158 pKa = 3.35DD159 pKa = 3.53TLARR163 pKa = 11.84MNSEE167 pKa = 3.7WVDD170 pKa = 3.68GYY172 pKa = 11.34LRR174 pKa = 11.84EE175 pKa = 4.04LAKK178 pKa = 10.08TGCIVKK184 pKa = 10.05LAEE187 pKa = 3.91NSRR190 pKa = 11.84EE191 pKa = 3.73FSGFRR196 pKa = 11.84VEE198 pKa = 5.06GDD200 pKa = 3.5SVANAIVTPIYY211 pKa = 9.75PEE213 pKa = 3.61KK214 pKa = 10.78HH215 pKa = 5.54KK216 pKa = 10.88FLVKK220 pKa = 10.31HH221 pKa = 5.46VRR223 pKa = 11.84AEE225 pKa = 4.2DD226 pKa = 3.46EE227 pKa = 4.24HH228 pKa = 8.63SVFLSFSLLYY238 pKa = 10.92ALARR242 pKa = 11.84PGWLNNVIGRR252 pKa = 11.84TDD254 pKa = 3.29VTVGPMQRR262 pKa = 11.84LWAKK266 pKa = 10.59GMAKK270 pKa = 10.45LEE272 pKa = 4.31LFDD275 pKa = 5.41TLPEE279 pKa = 3.89WTRR282 pKa = 11.84FWDD285 pKa = 3.36
MM1 pKa = 7.77VDD3 pKa = 3.23RR4 pKa = 11.84CLMHH8 pKa = 6.86PFIQSEE14 pKa = 4.03IRR16 pKa = 11.84NPMDD20 pKa = 3.43VVSKK24 pKa = 10.94AGWAPLPAGYY34 pKa = 10.28QRR36 pKa = 11.84MIAEE40 pKa = 4.38FPEE43 pKa = 4.18KK44 pKa = 9.67QAYY47 pKa = 10.11AIDD50 pKa = 3.75KK51 pKa = 10.41KK52 pKa = 10.9LWDD55 pKa = 3.4WTMPGWVVLEE65 pKa = 3.92YY66 pKa = 11.14VLAKK70 pKa = 10.35LRR72 pKa = 11.84QCRR75 pKa = 11.84EE76 pKa = 3.71HH77 pKa = 6.85SLLWEE82 pKa = 3.71WMVWRR87 pKa = 11.84RR88 pKa = 11.84IYY90 pKa = 11.16YY91 pKa = 9.86SFGPGAVLAEE101 pKa = 4.0PTGAIWRR108 pKa = 11.84QTFWGLMKK116 pKa = 10.57SGSLLTLSMNGASQYY131 pKa = 10.93LEE133 pKa = 4.8HH134 pKa = 6.87ALAWLRR140 pKa = 11.84MLDD143 pKa = 3.98GGPISWPPRR152 pKa = 11.84MWAMGDD158 pKa = 3.35DD159 pKa = 3.53TLARR163 pKa = 11.84MNSEE167 pKa = 3.7WVDD170 pKa = 3.68GYY172 pKa = 11.34LRR174 pKa = 11.84EE175 pKa = 4.04LAKK178 pKa = 10.08TGCIVKK184 pKa = 10.05LAEE187 pKa = 3.91NSRR190 pKa = 11.84EE191 pKa = 3.73FSGFRR196 pKa = 11.84VEE198 pKa = 5.06GDD200 pKa = 3.5SVANAIVTPIYY211 pKa = 9.75PEE213 pKa = 3.61KK214 pKa = 10.78HH215 pKa = 5.54KK216 pKa = 10.88FLVKK220 pKa = 10.31HH221 pKa = 5.46VRR223 pKa = 11.84AEE225 pKa = 4.2DD226 pKa = 3.46EE227 pKa = 4.24HH228 pKa = 8.63SVFLSFSLLYY238 pKa = 10.92ALARR242 pKa = 11.84PGWLNNVIGRR252 pKa = 11.84TDD254 pKa = 3.29VTVGPMQRR262 pKa = 11.84LWAKK266 pKa = 10.59GMAKK270 pKa = 10.45LEE272 pKa = 4.31LFDD275 pKa = 5.41TLPEE279 pKa = 3.89WTRR282 pKa = 11.84FWDD285 pKa = 3.36
Molecular weight: 32.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
825 |
285 |
540 |
412.5 |
46.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.758 ± 0.351 | 0.606 ± 0.236 |
4.364 ± 0.105 | 7.758 ± 0.763 |
3.515 ± 0.182 | 6.788 ± 0.122 |
1.697 ± 0.216 | 2.909 ± 0.503 |
5.576 ± 0.537 | 9.212 ± 0.881 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
4.242 ± 0.355 | 2.545 ± 0.047 |
4.485 ± 0.598 | 3.636 ± 0.625 |
6.303 ± 0.192 | 9.091 ± 1.84 |
5.212 ± 0.716 | 7.515 ± 0.449 |
3.03 ± 1.553 | 3.758 ± 0.317 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
