
Sulfolobus ellipsoid virus 1
Taxonomy: Viruses; Ovaliviridae; Alphaovalivirus
Average proteome isoelectric point is 6.6
Get precalculated fractions of proteins
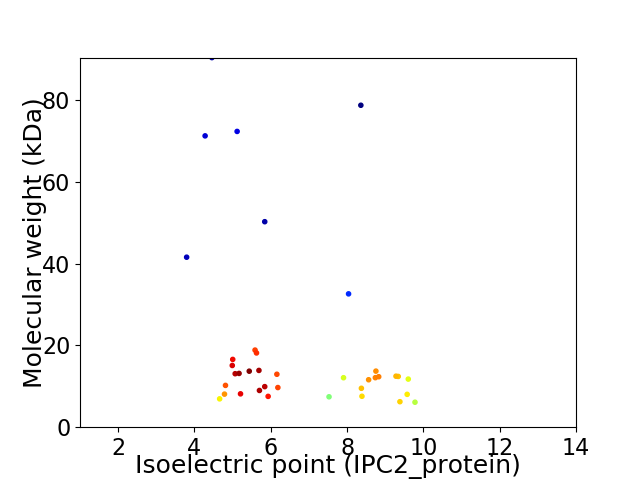
Virtual 2D-PAGE plot for 38 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2H4RBQ3|A0A2H4RBQ3_9VIRU Uncharacterized protein OS=Sulfolobus ellipsoid virus 1 OX=2056194 PE=4 SV=1
MM1 pKa = 7.47SFLSSLLKK9 pKa = 10.37ILLSPVIPIVPKK21 pKa = 10.22TPISTTTPVSTQTVTQLVTQTVTQVLTQYY50 pKa = 10.74EE51 pKa = 4.63YY52 pKa = 10.9VYY54 pKa = 10.76LSASCPVCPISTTVPQGYY72 pKa = 7.39TLVYY76 pKa = 9.6YY77 pKa = 8.44YY78 pKa = 10.4QISNPYY84 pKa = 9.62SQDD87 pKa = 3.34LNDD90 pKa = 3.3IQVPIEE96 pKa = 3.91LVVNNPLSYY105 pKa = 10.36IFCEE109 pKa = 4.09DD110 pKa = 3.39QACNKK115 pKa = 9.74PLFSWYY121 pKa = 9.56EE122 pKa = 3.51PSGIFWVLIPFIPANGDD139 pKa = 3.16IVIYY143 pKa = 10.07LYY145 pKa = 11.17EE146 pKa = 5.46NGDD149 pKa = 3.09VNYY152 pKa = 9.35PYY154 pKa = 10.12TGTFGSPLYY163 pKa = 10.85DD164 pKa = 3.18NGRR167 pKa = 11.84GVFYY171 pKa = 10.05EE172 pKa = 4.07YY173 pKa = 10.88FSFYY177 pKa = 10.18DD178 pKa = 3.72TLPHH182 pKa = 6.48EE183 pKa = 4.58WSTCYY188 pKa = 10.51NAVSPNYY195 pKa = 9.37NSTYY199 pKa = 9.8ATLPVNPPDD208 pKa = 3.82GGIGSDD214 pKa = 3.47YY215 pKa = 10.3AYY217 pKa = 9.84PVIGYY222 pKa = 8.71WIDD225 pKa = 3.69FYY227 pKa = 11.86VALPSTSSSDD237 pKa = 2.97ISSSVMALGVGSCNTTNTYY256 pKa = 10.57FIGGGGGDD264 pKa = 3.3TCGNNANGFAFCYY277 pKa = 10.72DD278 pKa = 3.21NFGTTNQWGVYY289 pKa = 9.87VGTTQVAPSPLNYY302 pKa = 9.36TPNNFALYY310 pKa = 10.15SIGVLSSSIIFQMNYY325 pKa = 9.34NNQFIYY331 pKa = 10.5SGSPPISMGNLVILDD346 pKa = 4.08SIGTIGLGFVRR357 pKa = 11.84MRR359 pKa = 11.84KK360 pKa = 8.11ATPDD364 pKa = 3.53GNNLPVLTFTPSGALASS381 pKa = 3.91
MM1 pKa = 7.47SFLSSLLKK9 pKa = 10.37ILLSPVIPIVPKK21 pKa = 10.22TPISTTTPVSTQTVTQLVTQTVTQVLTQYY50 pKa = 10.74EE51 pKa = 4.63YY52 pKa = 10.9VYY54 pKa = 10.76LSASCPVCPISTTVPQGYY72 pKa = 7.39TLVYY76 pKa = 9.6YY77 pKa = 8.44YY78 pKa = 10.4QISNPYY84 pKa = 9.62SQDD87 pKa = 3.34LNDD90 pKa = 3.3IQVPIEE96 pKa = 3.91LVVNNPLSYY105 pKa = 10.36IFCEE109 pKa = 4.09DD110 pKa = 3.39QACNKK115 pKa = 9.74PLFSWYY121 pKa = 9.56EE122 pKa = 3.51PSGIFWVLIPFIPANGDD139 pKa = 3.16IVIYY143 pKa = 10.07LYY145 pKa = 11.17EE146 pKa = 5.46NGDD149 pKa = 3.09VNYY152 pKa = 9.35PYY154 pKa = 10.12TGTFGSPLYY163 pKa = 10.85DD164 pKa = 3.18NGRR167 pKa = 11.84GVFYY171 pKa = 10.05EE172 pKa = 4.07YY173 pKa = 10.88FSFYY177 pKa = 10.18DD178 pKa = 3.72TLPHH182 pKa = 6.48EE183 pKa = 4.58WSTCYY188 pKa = 10.51NAVSPNYY195 pKa = 9.37NSTYY199 pKa = 9.8ATLPVNPPDD208 pKa = 3.82GGIGSDD214 pKa = 3.47YY215 pKa = 10.3AYY217 pKa = 9.84PVIGYY222 pKa = 8.71WIDD225 pKa = 3.69FYY227 pKa = 11.86VALPSTSSSDD237 pKa = 2.97ISSSVMALGVGSCNTTNTYY256 pKa = 10.57FIGGGGGDD264 pKa = 3.3TCGNNANGFAFCYY277 pKa = 10.72DD278 pKa = 3.21NFGTTNQWGVYY289 pKa = 9.87VGTTQVAPSPLNYY302 pKa = 9.36TPNNFALYY310 pKa = 10.15SIGVLSSSIIFQMNYY325 pKa = 9.34NNQFIYY331 pKa = 10.5SGSPPISMGNLVILDD346 pKa = 4.08SIGTIGLGFVRR357 pKa = 11.84MRR359 pKa = 11.84KK360 pKa = 8.11ATPDD364 pKa = 3.53GNNLPVLTFTPSGALASS381 pKa = 3.91
Molecular weight: 41.54 kDa
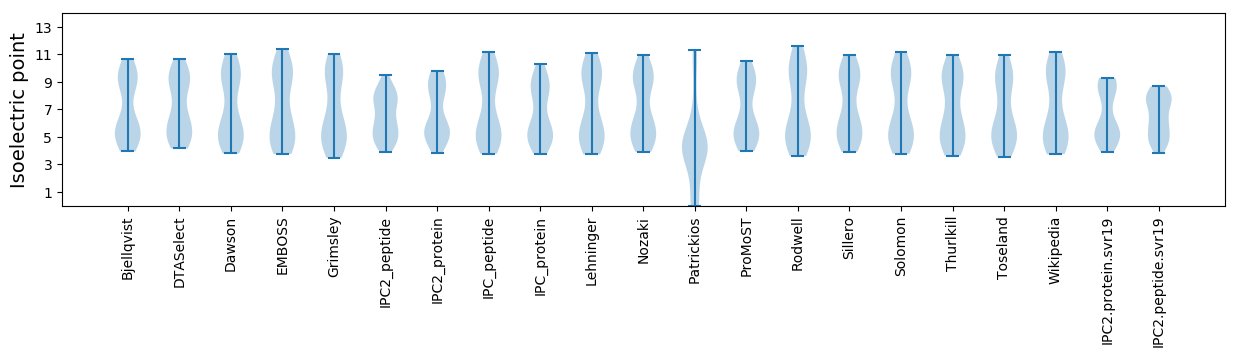
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2H4RBQ4|A0A2H4RBQ4_9VIRU Zn_C2H2-containing protein OS=Sulfolobus ellipsoid virus 1 OX=2056194 PE=4 SV=1
MM1 pKa = 7.68PKK3 pKa = 10.17ANRR6 pKa = 11.84YY7 pKa = 9.53AGLLTISLKK16 pKa = 10.7KK17 pKa = 10.68LLIEE21 pKa = 4.5LPIFDD26 pKa = 5.26RR27 pKa = 11.84LTLIKK32 pKa = 9.97IRR34 pKa = 11.84LISRR38 pKa = 11.84VIKK41 pKa = 9.92FHH43 pKa = 6.92SKK45 pKa = 10.73NNLCEE50 pKa = 4.4RR51 pKa = 11.84II52 pKa = 3.75
MM1 pKa = 7.68PKK3 pKa = 10.17ANRR6 pKa = 11.84YY7 pKa = 9.53AGLLTISLKK16 pKa = 10.7KK17 pKa = 10.68LLIEE21 pKa = 4.5LPIFDD26 pKa = 5.26RR27 pKa = 11.84LTLIKK32 pKa = 9.97IRR34 pKa = 11.84LISRR38 pKa = 11.84VIKK41 pKa = 9.92FHH43 pKa = 6.92SKK45 pKa = 10.73NNLCEE50 pKa = 4.4RR51 pKa = 11.84II52 pKa = 3.75
Molecular weight: 6.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6917 |
52 |
814 |
182.0 |
20.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.467 ± 0.245 | 1.084 ± 0.155 |
4.409 ± 0.421 | 4.901 ± 0.699 |
4.641 ± 0.328 | 5.523 ± 0.405 |
1.128 ± 0.198 | 9.122 ± 0.314 |
6.636 ± 1.047 | 9.441 ± 0.502 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.183 ± 0.26 | 7.417 ± 0.694 |
4.568 ± 0.452 | 3.354 ± 0.421 |
2.732 ± 0.467 | 7.619 ± 0.745 |
6.376 ± 0.589 | 6.954 ± 0.395 |
0.853 ± 0.093 | 6.592 ± 0.305 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
