
Vallicoccus soli
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Motilibacterales; Vallicoccaceae; Vallicoccus
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
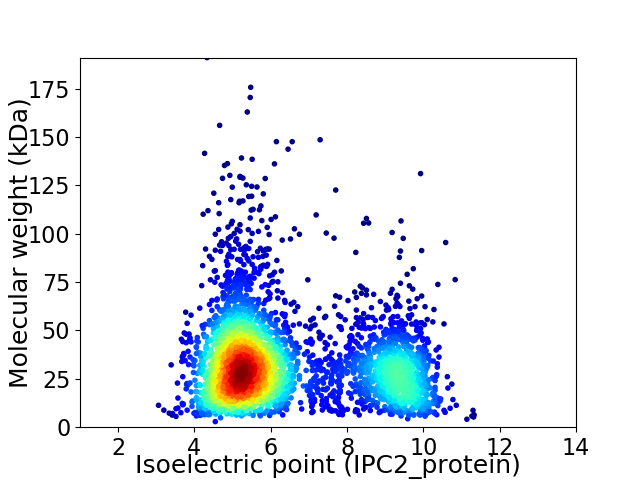
Virtual 2D-PAGE plot for 3658 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3A3ZCU1|A0A3A3ZCU1_9ACTN GAF domain-containing protein OS=Vallicoccus soli OX=2339232 GN=D5H78_18240 PE=4 SV=1
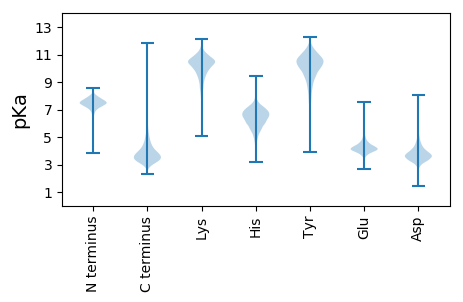
MM1 pKa = 7.72GSRR4 pKa = 11.84TNRR7 pKa = 11.84PAALAAGVLGLSLVLTACGSDD28 pKa = 2.94NGGDD32 pKa = 3.69DD33 pKa = 3.72TASGGSGGGSGADD46 pKa = 3.16CSAYY50 pKa = 9.76EE51 pKa = 4.84QYY53 pKa = 9.6TQDD56 pKa = 3.98FPGIDD61 pKa = 3.38GKK63 pKa = 8.7TVSVYY68 pKa = 10.13TSIVTPEE75 pKa = 4.34DD76 pKa = 3.53QPHH79 pKa = 6.74LDD81 pKa = 3.15SYY83 pKa = 11.92AAFEE87 pKa = 4.43EE88 pKa = 4.86CTGVDD93 pKa = 3.37VQYY96 pKa = 11.04EE97 pKa = 4.03GDD99 pKa = 3.56KK100 pKa = 11.34AFEE103 pKa = 4.06AQVLVRR109 pKa = 11.84AKK111 pKa = 10.57AGNAPDD117 pKa = 3.46IAYY120 pKa = 9.95VPQPGLLQQLVATGAAVPAPEE141 pKa = 3.72QTAANVDD148 pKa = 3.76EE149 pKa = 5.69FFGEE153 pKa = 4.03DD154 pKa = 2.72WKK156 pKa = 11.46GYY158 pKa = 10.47GSVDD162 pKa = 3.33GQFYY166 pKa = 10.39AAPLGANVKK175 pKa = 10.29SFVWYY180 pKa = 10.25SPSEE184 pKa = 4.2FEE186 pKa = 4.42DD187 pKa = 3.08SGYY190 pKa = 10.79EE191 pKa = 4.01VPTTLDD197 pKa = 3.43EE198 pKa = 4.81LKK200 pKa = 10.88SLTDD204 pKa = 3.38QMVADD209 pKa = 5.65GKK211 pKa = 10.73VPWCAGIASGEE222 pKa = 4.07ATGWPVTDD230 pKa = 3.15WMEE233 pKa = 5.81DD234 pKa = 2.78MMLRR238 pKa = 11.84LNGPEE243 pKa = 5.81VYY245 pKa = 10.11DD246 pKa = 2.92QWVNHH251 pKa = 6.9EE252 pKa = 4.35IPFNGPEE259 pKa = 3.82STAALDD265 pKa = 4.35AVGEE269 pKa = 4.11YY270 pKa = 10.41LKK272 pKa = 11.08NPEE275 pKa = 4.41YY276 pKa = 11.46VNGGYY281 pKa = 11.09GDD283 pKa = 3.7VTTIATTTFQDD294 pKa = 3.06GGLPIIDD301 pKa = 4.33GACSLHH307 pKa = 6.07RR308 pKa = 11.84QAGFYY313 pKa = 9.29AANWPEE319 pKa = 3.99GTEE322 pKa = 3.98VAEE325 pKa = 5.08DD326 pKa = 3.8GDD328 pKa = 3.98VFAFYY333 pKa = 10.95LPGQTAEE340 pKa = 4.41DD341 pKa = 4.06RR342 pKa = 11.84PVLGGGEE349 pKa = 4.04FVLAFDD355 pKa = 5.84DD356 pKa = 4.14RR357 pKa = 11.84PEE359 pKa = 3.96VQAFQTFLSSDD370 pKa = 2.5TWANEE375 pKa = 3.67KK376 pKa = 10.68AIATPQGGWVSANSGLEE393 pKa = 3.67VDD395 pKa = 4.66NLVSPIDD402 pKa = 3.62QLSAEE407 pKa = 4.47ILQDD411 pKa = 3.4PNAVFRR417 pKa = 11.84FDD419 pKa = 5.93GSDD422 pKa = 3.44QMPGAVGADD431 pKa = 3.73AFWKK435 pKa = 10.38QATNWITGQSTQEE448 pKa = 4.02TVDD451 pKa = 3.93KK452 pKa = 11.25IEE454 pKa = 4.4AAWPASS460 pKa = 3.41
MM1 pKa = 7.72GSRR4 pKa = 11.84TNRR7 pKa = 11.84PAALAAGVLGLSLVLTACGSDD28 pKa = 2.94NGGDD32 pKa = 3.69DD33 pKa = 3.72TASGGSGGGSGADD46 pKa = 3.16CSAYY50 pKa = 9.76EE51 pKa = 4.84QYY53 pKa = 9.6TQDD56 pKa = 3.98FPGIDD61 pKa = 3.38GKK63 pKa = 8.7TVSVYY68 pKa = 10.13TSIVTPEE75 pKa = 4.34DD76 pKa = 3.53QPHH79 pKa = 6.74LDD81 pKa = 3.15SYY83 pKa = 11.92AAFEE87 pKa = 4.43EE88 pKa = 4.86CTGVDD93 pKa = 3.37VQYY96 pKa = 11.04EE97 pKa = 4.03GDD99 pKa = 3.56KK100 pKa = 11.34AFEE103 pKa = 4.06AQVLVRR109 pKa = 11.84AKK111 pKa = 10.57AGNAPDD117 pKa = 3.46IAYY120 pKa = 9.95VPQPGLLQQLVATGAAVPAPEE141 pKa = 3.72QTAANVDD148 pKa = 3.76EE149 pKa = 5.69FFGEE153 pKa = 4.03DD154 pKa = 2.72WKK156 pKa = 11.46GYY158 pKa = 10.47GSVDD162 pKa = 3.33GQFYY166 pKa = 10.39AAPLGANVKK175 pKa = 10.29SFVWYY180 pKa = 10.25SPSEE184 pKa = 4.2FEE186 pKa = 4.42DD187 pKa = 3.08SGYY190 pKa = 10.79EE191 pKa = 4.01VPTTLDD197 pKa = 3.43EE198 pKa = 4.81LKK200 pKa = 10.88SLTDD204 pKa = 3.38QMVADD209 pKa = 5.65GKK211 pKa = 10.73VPWCAGIASGEE222 pKa = 4.07ATGWPVTDD230 pKa = 3.15WMEE233 pKa = 5.81DD234 pKa = 2.78MMLRR238 pKa = 11.84LNGPEE243 pKa = 5.81VYY245 pKa = 10.11DD246 pKa = 2.92QWVNHH251 pKa = 6.9EE252 pKa = 4.35IPFNGPEE259 pKa = 3.82STAALDD265 pKa = 4.35AVGEE269 pKa = 4.11YY270 pKa = 10.41LKK272 pKa = 11.08NPEE275 pKa = 4.41YY276 pKa = 11.46VNGGYY281 pKa = 11.09GDD283 pKa = 3.7VTTIATTTFQDD294 pKa = 3.06GGLPIIDD301 pKa = 4.33GACSLHH307 pKa = 6.07RR308 pKa = 11.84QAGFYY313 pKa = 9.29AANWPEE319 pKa = 3.99GTEE322 pKa = 3.98VAEE325 pKa = 5.08DD326 pKa = 3.8GDD328 pKa = 3.98VFAFYY333 pKa = 10.95LPGQTAEE340 pKa = 4.41DD341 pKa = 4.06RR342 pKa = 11.84PVLGGGEE349 pKa = 4.04FVLAFDD355 pKa = 5.84DD356 pKa = 4.14RR357 pKa = 11.84PEE359 pKa = 3.96VQAFQTFLSSDD370 pKa = 2.5TWANEE375 pKa = 3.67KK376 pKa = 10.68AIATPQGGWVSANSGLEE393 pKa = 3.67VDD395 pKa = 4.66NLVSPIDD402 pKa = 3.62QLSAEE407 pKa = 4.47ILQDD411 pKa = 3.4PNAVFRR417 pKa = 11.84FDD419 pKa = 5.93GSDD422 pKa = 3.44QMPGAVGADD431 pKa = 3.73AFWKK435 pKa = 10.38QATNWITGQSTQEE448 pKa = 4.02TVDD451 pKa = 3.93KK452 pKa = 11.25IEE454 pKa = 4.4AAWPASS460 pKa = 3.41
Molecular weight: 48.73 kDa
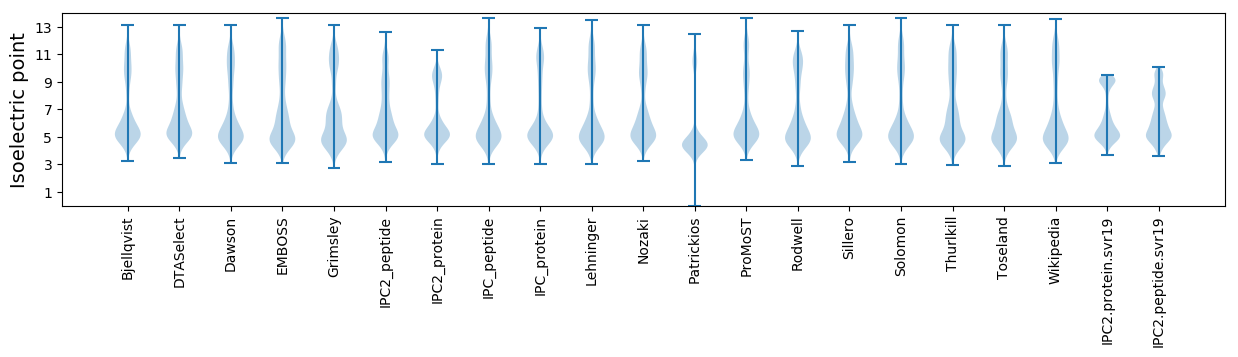
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3A3YTM2|A0A3A3YTM2_9ACTN Iron-sulfur protein OS=Vallicoccus soli OX=2339232 GN=D5H78_13320 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SKK15 pKa = 8.58THH17 pKa = 5.46GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.75GRR40 pKa = 11.84ASLSAA45 pKa = 3.83
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84SKK15 pKa = 8.58THH17 pKa = 5.46GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.75GRR40 pKa = 11.84ASLSAA45 pKa = 3.83
Molecular weight: 5.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1198445 |
29 |
1814 |
327.6 |
34.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
15.525 ± 0.077 | 0.688 ± 0.01 |
6.11 ± 0.03 | 5.652 ± 0.04 |
2.155 ± 0.027 | 10.35 ± 0.041 |
2.012 ± 0.021 | 1.698 ± 0.031 |
1.119 ± 0.021 | 11.139 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.336 ± 0.014 | 1.09 ± 0.018 |
6.506 ± 0.034 | 2.545 ± 0.021 |
9.159 ± 0.053 | 4.228 ± 0.03 |
5.177 ± 0.03 | 10.338 ± 0.037 |
1.433 ± 0.015 | 1.741 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
