
Moraxella atlantae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Moraxellaceae; Moraxella
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
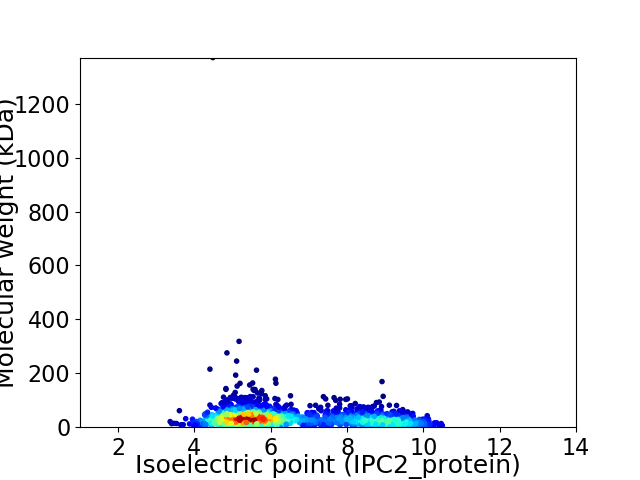
Virtual 2D-PAGE plot for 1904 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B8Q9F9|A0A1B8Q9F9_9GAMM Putative manganese efflux pump MntP OS=Moraxella atlantae OX=34059 GN=mntP PE=3 SV=1
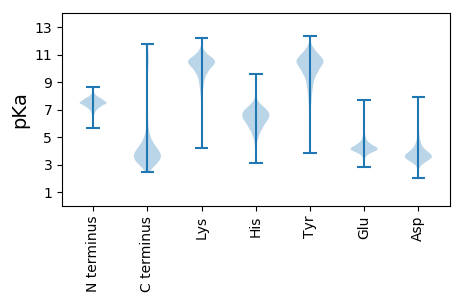
MM1 pKa = 7.25MNDD4 pKa = 3.11TMTLTDD10 pKa = 3.26NAANKK15 pKa = 9.12VRR17 pKa = 11.84RR18 pKa = 11.84LRR20 pKa = 11.84AQEE23 pKa = 4.03GNEE26 pKa = 3.74DD27 pKa = 3.5LMLRR31 pKa = 11.84VYY33 pKa = 9.16VTGGGCSGFSYY44 pKa = 10.51GFNFEE49 pKa = 4.19EE50 pKa = 5.03QLADD54 pKa = 5.2DD55 pKa = 5.16DD56 pKa = 4.7AQFANGDD63 pKa = 3.5VALVVDD69 pKa = 4.29SLSYY73 pKa = 10.65QYY75 pKa = 11.78LHH77 pKa = 7.14GSTIDD82 pKa = 3.68YY83 pKa = 10.87IEE85 pKa = 4.37GLEE88 pKa = 4.09GSRR91 pKa = 11.84FVVDD95 pKa = 3.95NPNATTTCGCGSSFSII111 pKa = 5.02
MM1 pKa = 7.25MNDD4 pKa = 3.11TMTLTDD10 pKa = 3.26NAANKK15 pKa = 9.12VRR17 pKa = 11.84RR18 pKa = 11.84LRR20 pKa = 11.84AQEE23 pKa = 4.03GNEE26 pKa = 3.74DD27 pKa = 3.5LMLRR31 pKa = 11.84VYY33 pKa = 9.16VTGGGCSGFSYY44 pKa = 10.51GFNFEE49 pKa = 4.19EE50 pKa = 5.03QLADD54 pKa = 5.2DD55 pKa = 5.16DD56 pKa = 4.7AQFANGDD63 pKa = 3.5VALVVDD69 pKa = 4.29SLSYY73 pKa = 10.65QYY75 pKa = 11.78LHH77 pKa = 7.14GSTIDD82 pKa = 3.68YY83 pKa = 10.87IEE85 pKa = 4.37GLEE88 pKa = 4.09GSRR91 pKa = 11.84FVVDD95 pKa = 3.95NPNATTTCGCGSSFSII111 pKa = 5.02
Molecular weight: 12.04 kDa
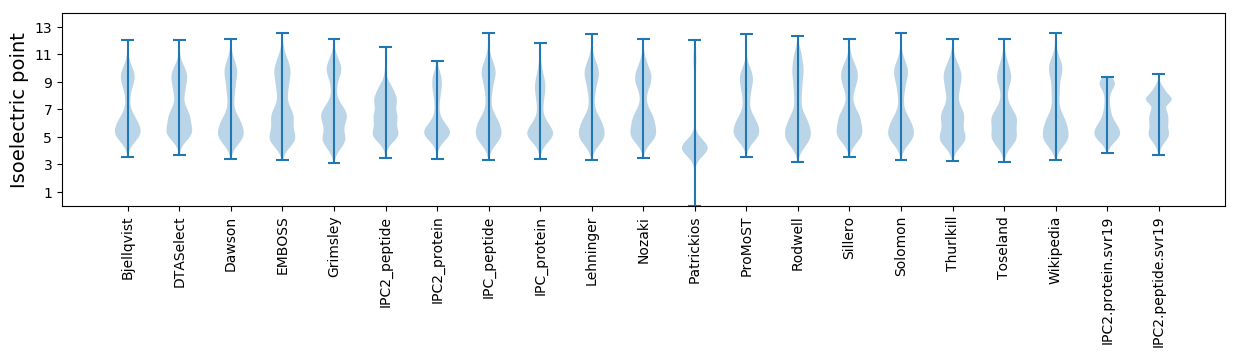
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B8QDT3|A0A1B8QDT3_9GAMM Rep_3 domain-containing protein OS=Moraxella atlantae OX=34059 GN=A9306_08355 PE=4 SV=1
MM1 pKa = 7.62LGLFCRR7 pKa = 11.84GKK9 pKa = 9.68WFTVKK14 pKa = 10.53NPAQHH19 pKa = 6.59KK20 pKa = 8.61VFIMPKK26 pKa = 9.06HH27 pKa = 5.69QATQTDD33 pKa = 4.94RR34 pKa = 11.84ITTSDD39 pKa = 3.47TPQVTRR45 pKa = 11.84RR46 pKa = 11.84CLRR49 pKa = 11.84RR50 pKa = 11.84ARR52 pKa = 11.84RR53 pKa = 11.84VLSSAEE59 pKa = 3.8RR60 pKa = 11.84RR61 pKa = 11.84HH62 pKa = 5.96AAGQAVRR69 pKa = 11.84RR70 pKa = 11.84LSRR73 pKa = 11.84LPLPRR78 pKa = 11.84QPNRR82 pKa = 11.84PLNIGVYY89 pKa = 10.27ADD91 pKa = 4.29AFGEE95 pKa = 4.42LPTQGLIDD103 pKa = 3.35WARR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84GWRR111 pKa = 11.84VFLPVVIHH119 pKa = 6.35AHH121 pKa = 5.42SALCFRR127 pKa = 11.84QLHH130 pKa = 5.39ATTLKK135 pKa = 9.16NARR138 pKa = 11.84LARR141 pKa = 11.84HH142 pKa = 6.14PLAMRR147 pKa = 11.84QPAYY151 pKa = 10.28GALVTARR158 pKa = 11.84DD159 pKa = 3.81LDD161 pKa = 4.0VLFVPLVAFDD171 pKa = 4.26DD172 pKa = 4.17DD173 pKa = 4.76GYY175 pKa = 11.06RR176 pKa = 11.84LGMGGGFYY184 pKa = 10.82DD185 pKa = 3.01RR186 pKa = 11.84TLAHH190 pKa = 6.92RR191 pKa = 11.84RR192 pKa = 11.84NKK194 pKa = 10.05PLVIGWAYY202 pKa = 9.64DD203 pKa = 3.53WQRR206 pKa = 11.84VAHH209 pKa = 7.03LPRR212 pKa = 11.84QPWDD216 pKa = 3.19ARR218 pKa = 11.84LDD220 pKa = 3.96LAVLPSGIMDD230 pKa = 3.65FRR232 pKa = 11.84QRR234 pKa = 11.84FVVSPPKK241 pKa = 10.26PVTAAPNDD249 pKa = 3.76FLLKK253 pKa = 10.71LLTCSTPTVANDD265 pKa = 3.33AAVFDD270 pKa = 5.33DD271 pKa = 5.71DD272 pKa = 4.62VAPVDD277 pKa = 4.2DD278 pKa = 4.74TLSSAQIVTVMNPNHH293 pKa = 6.98LAQLLAQADD302 pKa = 3.49AFLAVNRR309 pKa = 11.84DD310 pKa = 3.19TDD312 pKa = 3.57SDD314 pKa = 3.93FF315 pKa = 4.7
MM1 pKa = 7.62LGLFCRR7 pKa = 11.84GKK9 pKa = 9.68WFTVKK14 pKa = 10.53NPAQHH19 pKa = 6.59KK20 pKa = 8.61VFIMPKK26 pKa = 9.06HH27 pKa = 5.69QATQTDD33 pKa = 4.94RR34 pKa = 11.84ITTSDD39 pKa = 3.47TPQVTRR45 pKa = 11.84RR46 pKa = 11.84CLRR49 pKa = 11.84RR50 pKa = 11.84ARR52 pKa = 11.84RR53 pKa = 11.84VLSSAEE59 pKa = 3.8RR60 pKa = 11.84RR61 pKa = 11.84HH62 pKa = 5.96AAGQAVRR69 pKa = 11.84RR70 pKa = 11.84LSRR73 pKa = 11.84LPLPRR78 pKa = 11.84QPNRR82 pKa = 11.84PLNIGVYY89 pKa = 10.27ADD91 pKa = 4.29AFGEE95 pKa = 4.42LPTQGLIDD103 pKa = 3.35WARR106 pKa = 11.84RR107 pKa = 11.84RR108 pKa = 11.84GWRR111 pKa = 11.84VFLPVVIHH119 pKa = 6.35AHH121 pKa = 5.42SALCFRR127 pKa = 11.84QLHH130 pKa = 5.39ATTLKK135 pKa = 9.16NARR138 pKa = 11.84LARR141 pKa = 11.84HH142 pKa = 6.14PLAMRR147 pKa = 11.84QPAYY151 pKa = 10.28GALVTARR158 pKa = 11.84DD159 pKa = 3.81LDD161 pKa = 4.0VLFVPLVAFDD171 pKa = 4.26DD172 pKa = 4.17DD173 pKa = 4.76GYY175 pKa = 11.06RR176 pKa = 11.84LGMGGGFYY184 pKa = 10.82DD185 pKa = 3.01RR186 pKa = 11.84TLAHH190 pKa = 6.92RR191 pKa = 11.84RR192 pKa = 11.84NKK194 pKa = 10.05PLVIGWAYY202 pKa = 9.64DD203 pKa = 3.53WQRR206 pKa = 11.84VAHH209 pKa = 7.03LPRR212 pKa = 11.84QPWDD216 pKa = 3.19ARR218 pKa = 11.84LDD220 pKa = 3.96LAVLPSGIMDD230 pKa = 3.65FRR232 pKa = 11.84QRR234 pKa = 11.84FVVSPPKK241 pKa = 10.26PVTAAPNDD249 pKa = 3.76FLLKK253 pKa = 10.71LLTCSTPTVANDD265 pKa = 3.33AAVFDD270 pKa = 5.33DD271 pKa = 5.71DD272 pKa = 4.62VAPVDD277 pKa = 4.2DD278 pKa = 4.74TLSSAQIVTVMNPNHH293 pKa = 6.98LAQLLAQADD302 pKa = 3.49AFLAVNRR309 pKa = 11.84DD310 pKa = 3.19TDD312 pKa = 3.57SDD314 pKa = 3.93FF315 pKa = 4.7
Molecular weight: 35.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
631180 |
38 |
12592 |
331.5 |
36.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.696 ± 0.084 | 0.876 ± 0.023 |
6.08 ± 0.053 | 4.521 ± 0.055 |
3.576 ± 0.045 | 6.584 ± 0.058 |
2.422 ± 0.039 | 5.706 ± 0.039 |
4.385 ± 0.05 | 10.428 ± 0.075 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.407 ± 0.033 | 4.317 ± 0.067 |
4.416 ± 0.037 | 5.601 ± 0.056 |
5.031 ± 0.052 | 5.5 ± 0.056 |
6.172 ± 0.073 | 7.045 ± 0.054 |
1.216 ± 0.024 | 3.018 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
