
Chickpea chlorotic dwarf virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Geminiviridae; Mastrevirus
Average proteome isoelectric point is 7.01
Get precalculated fractions of proteins
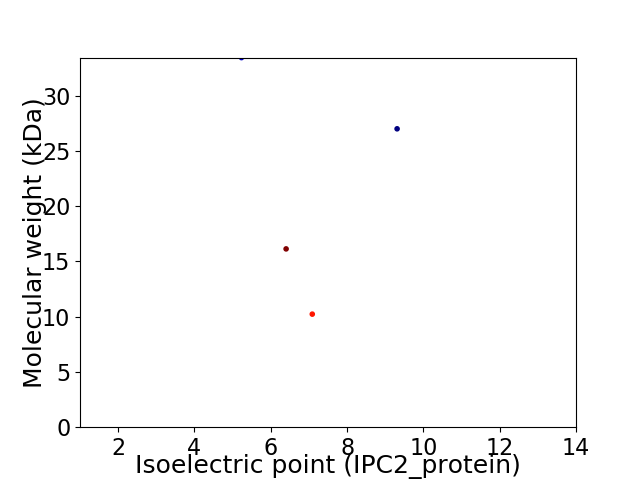
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F8K7V6|F8K7V6_9GEMI Replication-associated protein OS=Chickpea chlorotic dwarf virus OX=463360 GN=repA PE=3 SV=1
MM1 pKa = 7.73PSANKK6 pKa = 9.92NFRR9 pKa = 11.84FQSKK13 pKa = 8.6YY14 pKa = 10.68VFLTYY19 pKa = 9.4PKK21 pKa = 10.35CSSQRR26 pKa = 11.84DD27 pKa = 3.55ALLEE31 pKa = 4.0FLWEE35 pKa = 4.03KK36 pKa = 9.44LTPFLIYY43 pKa = 10.06FIGVATEE50 pKa = 3.71LHH52 pKa = 5.95QDD54 pKa = 3.29GTTHH58 pKa = 5.38YY59 pKa = 9.89HH60 pKa = 7.28ALIQLDD66 pKa = 4.12KK67 pKa = 10.29RR68 pKa = 11.84PHH70 pKa = 6.03IRR72 pKa = 11.84DD73 pKa = 3.11PSFFDD78 pKa = 4.7FEE80 pKa = 4.9GNHH83 pKa = 6.71PNIQPARR90 pKa = 11.84NSKK93 pKa = 10.03QVLDD97 pKa = 4.6YY98 pKa = 10.62ISKK101 pKa = 11.02DD102 pKa = 3.01GDD104 pKa = 3.15IKK106 pKa = 10.39TRR108 pKa = 11.84GDD110 pKa = 3.45FRR112 pKa = 11.84DD113 pKa = 3.79HH114 pKa = 7.01KK115 pKa = 10.4ISPSKK120 pKa = 10.85SDD122 pKa = 3.27ARR124 pKa = 11.84WRR126 pKa = 11.84TIIQTATTKK135 pKa = 10.79EE136 pKa = 4.21EE137 pKa = 3.97YY138 pKa = 10.81LDD140 pKa = 3.86MIKK143 pKa = 10.68EE144 pKa = 4.22EE145 pKa = 5.22FPHH148 pKa = 6.19EE149 pKa = 4.13WATKK153 pKa = 8.91LQWLEE158 pKa = 3.62YY159 pKa = 9.89SANKK163 pKa = 9.86LFPPQPEE170 pKa = 4.52PYY172 pKa = 9.63VSPFTEE178 pKa = 3.78SDD180 pKa = 3.68LRR182 pKa = 11.84CHH184 pKa = 6.88EE185 pKa = 5.67DD186 pKa = 2.87LAAWRR191 pKa = 11.84DD192 pKa = 3.17KK193 pKa = 11.02HH194 pKa = 6.79LYY196 pKa = 9.05HH197 pKa = 7.98VSIDD201 pKa = 3.24AYY203 pKa = 9.26TYY205 pKa = 8.99IHH207 pKa = 5.86TVSYY211 pKa = 9.8EE212 pKa = 3.98QAEE215 pKa = 4.28SDD217 pKa = 4.41LDD219 pKa = 3.52WMADD223 pKa = 3.35LTRR226 pKa = 11.84TMEE229 pKa = 4.45GLGSDD234 pKa = 4.04TPVSTSADD242 pKa = 3.27QLVPEE247 pKa = 5.18RR248 pKa = 11.84PPGLEE253 pKa = 3.87VSDD256 pKa = 4.08DD257 pKa = 3.7TTTGMVPSISQRR269 pKa = 11.84TMTMPPTTSSTTSPSSSSHH288 pKa = 6.34CGSSS292 pKa = 2.91
MM1 pKa = 7.73PSANKK6 pKa = 9.92NFRR9 pKa = 11.84FQSKK13 pKa = 8.6YY14 pKa = 10.68VFLTYY19 pKa = 9.4PKK21 pKa = 10.35CSSQRR26 pKa = 11.84DD27 pKa = 3.55ALLEE31 pKa = 4.0FLWEE35 pKa = 4.03KK36 pKa = 9.44LTPFLIYY43 pKa = 10.06FIGVATEE50 pKa = 3.71LHH52 pKa = 5.95QDD54 pKa = 3.29GTTHH58 pKa = 5.38YY59 pKa = 9.89HH60 pKa = 7.28ALIQLDD66 pKa = 4.12KK67 pKa = 10.29RR68 pKa = 11.84PHH70 pKa = 6.03IRR72 pKa = 11.84DD73 pKa = 3.11PSFFDD78 pKa = 4.7FEE80 pKa = 4.9GNHH83 pKa = 6.71PNIQPARR90 pKa = 11.84NSKK93 pKa = 10.03QVLDD97 pKa = 4.6YY98 pKa = 10.62ISKK101 pKa = 11.02DD102 pKa = 3.01GDD104 pKa = 3.15IKK106 pKa = 10.39TRR108 pKa = 11.84GDD110 pKa = 3.45FRR112 pKa = 11.84DD113 pKa = 3.79HH114 pKa = 7.01KK115 pKa = 10.4ISPSKK120 pKa = 10.85SDD122 pKa = 3.27ARR124 pKa = 11.84WRR126 pKa = 11.84TIIQTATTKK135 pKa = 10.79EE136 pKa = 4.21EE137 pKa = 3.97YY138 pKa = 10.81LDD140 pKa = 3.86MIKK143 pKa = 10.68EE144 pKa = 4.22EE145 pKa = 5.22FPHH148 pKa = 6.19EE149 pKa = 4.13WATKK153 pKa = 8.91LQWLEE158 pKa = 3.62YY159 pKa = 9.89SANKK163 pKa = 9.86LFPPQPEE170 pKa = 4.52PYY172 pKa = 9.63VSPFTEE178 pKa = 3.78SDD180 pKa = 3.68LRR182 pKa = 11.84CHH184 pKa = 6.88EE185 pKa = 5.67DD186 pKa = 2.87LAAWRR191 pKa = 11.84DD192 pKa = 3.17KK193 pKa = 11.02HH194 pKa = 6.79LYY196 pKa = 9.05HH197 pKa = 7.98VSIDD201 pKa = 3.24AYY203 pKa = 9.26TYY205 pKa = 8.99IHH207 pKa = 5.86TVSYY211 pKa = 9.8EE212 pKa = 3.98QAEE215 pKa = 4.28SDD217 pKa = 4.41LDD219 pKa = 3.52WMADD223 pKa = 3.35LTRR226 pKa = 11.84TMEE229 pKa = 4.45GLGSDD234 pKa = 4.04TPVSTSADD242 pKa = 3.27QLVPEE247 pKa = 5.18RR248 pKa = 11.84PPGLEE253 pKa = 3.87VSDD256 pKa = 4.08DD257 pKa = 3.7TTTGMVPSISQRR269 pKa = 11.84TMTMPPTTSSTTSPSSSSHH288 pKa = 6.34CGSSS292 pKa = 2.91
Molecular weight: 33.45 kDa
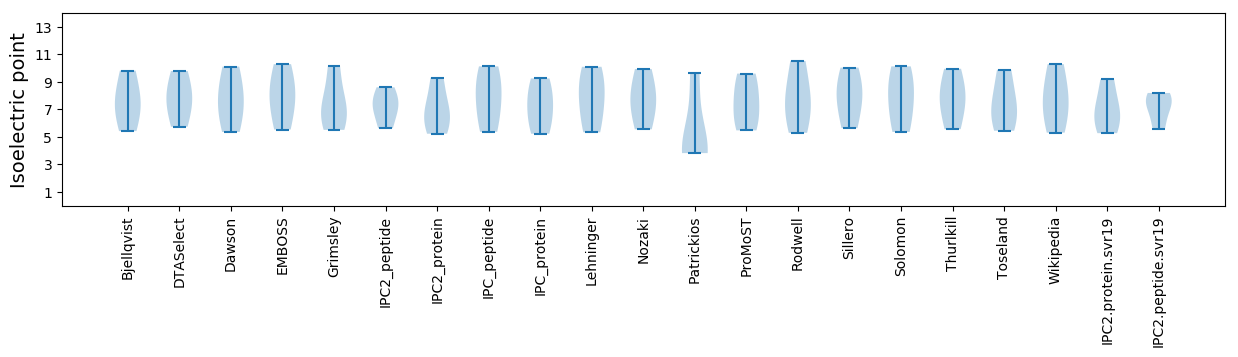
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F8K7V5|F8K7V5_9GEMI Replication-associated protein (Fragment) OS=Chickpea chlorotic dwarf virus OX=463360 GN=repB PE=4 SV=1
MM1 pKa = 7.12STSTWGKK8 pKa = 8.27KK9 pKa = 8.03RR10 pKa = 11.84KK11 pKa = 9.59RR12 pKa = 11.84SGAEE16 pKa = 3.43SRR18 pKa = 11.84AKK20 pKa = 10.38SRR22 pKa = 11.84ASGSYY27 pKa = 9.01VPRR30 pKa = 11.84SVTSRR35 pKa = 11.84RR36 pKa = 11.84EE37 pKa = 3.96SLQVATFSWTSSGSGIKK54 pKa = 10.49FSAGGAAYY62 pKa = 9.97LVSNFPQGANDD73 pKa = 3.63NCRR76 pKa = 11.84HH77 pKa = 4.98TNKK80 pKa = 9.3TVLYY84 pKa = 10.59KK85 pKa = 10.37FMAKK89 pKa = 8.15NTIYY93 pKa = 10.54LDD95 pKa = 3.32SSHH98 pKa = 6.16YY99 pKa = 10.86SKK101 pKa = 11.11VFRR104 pKa = 11.84CPFTFWLVYY113 pKa = 10.72DD114 pKa = 4.55KK115 pKa = 11.51APGANVPSTSDD126 pKa = 2.85IFEE129 pKa = 4.59GPSLFPNNPWTWTVSRR145 pKa = 11.84AACHH149 pKa = 5.68RR150 pKa = 11.84FVVKK154 pKa = 9.21KK155 pKa = 8.29TWSCTVEE162 pKa = 4.23SNGIDD167 pKa = 3.36PGKK170 pKa = 10.57AQGSTYY176 pKa = 10.37YY177 pKa = 11.11GPGPCNQVKK186 pKa = 10.24SCNKK190 pKa = 8.07FFKK193 pKa = 10.55RR194 pKa = 11.84LGVSTEE200 pKa = 3.87WKK202 pKa = 10.02NSSTGDD208 pKa = 3.25VGDD211 pKa = 3.7IKK213 pKa = 10.92EE214 pKa = 4.06GALYY218 pKa = 10.17IVGAPSQKK226 pKa = 10.34SDD228 pKa = 3.54VYY230 pKa = 9.4VNGYY234 pKa = 8.88FRR236 pKa = 11.84VYY238 pKa = 10.14FKK240 pKa = 11.17SVGNQQ245 pKa = 2.8
MM1 pKa = 7.12STSTWGKK8 pKa = 8.27KK9 pKa = 8.03RR10 pKa = 11.84KK11 pKa = 9.59RR12 pKa = 11.84SGAEE16 pKa = 3.43SRR18 pKa = 11.84AKK20 pKa = 10.38SRR22 pKa = 11.84ASGSYY27 pKa = 9.01VPRR30 pKa = 11.84SVTSRR35 pKa = 11.84RR36 pKa = 11.84EE37 pKa = 3.96SLQVATFSWTSSGSGIKK54 pKa = 10.49FSAGGAAYY62 pKa = 9.97LVSNFPQGANDD73 pKa = 3.63NCRR76 pKa = 11.84HH77 pKa = 4.98TNKK80 pKa = 9.3TVLYY84 pKa = 10.59KK85 pKa = 10.37FMAKK89 pKa = 8.15NTIYY93 pKa = 10.54LDD95 pKa = 3.32SSHH98 pKa = 6.16YY99 pKa = 10.86SKK101 pKa = 11.11VFRR104 pKa = 11.84CPFTFWLVYY113 pKa = 10.72DD114 pKa = 4.55KK115 pKa = 11.51APGANVPSTSDD126 pKa = 2.85IFEE129 pKa = 4.59GPSLFPNNPWTWTVSRR145 pKa = 11.84AACHH149 pKa = 5.68RR150 pKa = 11.84FVVKK154 pKa = 9.21KK155 pKa = 8.29TWSCTVEE162 pKa = 4.23SNGIDD167 pKa = 3.36PGKK170 pKa = 10.57AQGSTYY176 pKa = 10.37YY177 pKa = 11.11GPGPCNQVKK186 pKa = 10.24SCNKK190 pKa = 8.07FFKK193 pKa = 10.55RR194 pKa = 11.84LGVSTEE200 pKa = 3.87WKK202 pKa = 10.02NSSTGDD208 pKa = 3.25VGDD211 pKa = 3.7IKK213 pKa = 10.92EE214 pKa = 4.06GALYY218 pKa = 10.17IVGAPSQKK226 pKa = 10.34SDD228 pKa = 3.54VYY230 pKa = 9.4VNGYY234 pKa = 8.88FRR236 pKa = 11.84VYY238 pKa = 10.14FKK240 pKa = 11.17SVGNQQ245 pKa = 2.8
Molecular weight: 27.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
769 |
92 |
292 |
192.3 |
21.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.072 ± 0.789 | 1.821 ± 0.446 |
6.372 ± 1.125 | 3.771 ± 1.022 |
5.202 ± 0.345 | 6.112 ± 1.393 |
2.731 ± 0.68 | 4.681 ± 1.088 |
6.372 ± 0.795 | 5.722 ± 1.156 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.56 ± 0.368 | 4.031 ± 0.976 |
6.502 ± 0.732 | 3.771 ± 0.606 |
4.811 ± 0.205 | 10.663 ± 1.177 |
8.062 ± 0.599 | 5.852 ± 1.613 |
2.211 ± 0.427 | 4.681 ± 0.338 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
