
Wohlfahrtiimonas chitiniclastica SH04
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Gammaproteobacteria incertae sedis; Wohlfahrtiimonas; Wohlfahrtiimonas chitiniclastica
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
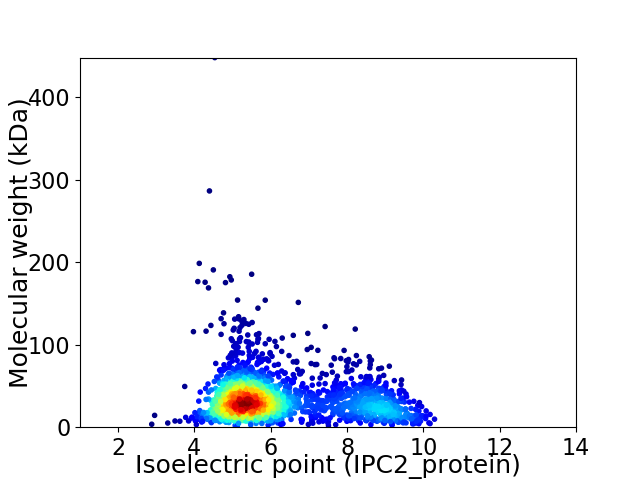
Virtual 2D-PAGE plot for 2004 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L8XUV4|L8XUV4_9GAMM Uncharacterized protein OS=Wohlfahrtiimonas chitiniclastica SH04 OX=1261130 GN=F387_01496 PE=4 SV=1
MM1 pKa = 7.55TYY3 pKa = 10.67KK4 pKa = 9.54KK5 pKa = 8.29TALSIASLLILGTFSYY21 pKa = 10.61AQQTPTEE28 pKa = 4.44LEE30 pKa = 3.97PNGIIAYY37 pKa = 9.15FAEE40 pKa = 5.6DD41 pKa = 3.51GTLLPSRR48 pKa = 11.84EE49 pKa = 3.43GSEE52 pKa = 4.5YY53 pKa = 10.03YY54 pKa = 10.27RR55 pKa = 11.84HH56 pKa = 6.13FKK58 pKa = 10.89DD59 pKa = 4.05VVSGCYY65 pKa = 9.46LVQDD69 pKa = 5.43FYY71 pKa = 11.99SQNDD75 pKa = 3.44QKK77 pKa = 10.24QTDD80 pKa = 5.09PICFMDD86 pKa = 4.54PGEE89 pKa = 4.13LQSWFPQSLYY99 pKa = 11.12GPLTFWHH106 pKa = 6.98KK107 pKa = 11.04DD108 pKa = 3.01GTKK111 pKa = 9.96AQSGYY116 pKa = 9.12SQQDD120 pKa = 3.05GGGNGTWTTWDD131 pKa = 3.23QDD133 pKa = 3.85GNASDD138 pKa = 5.29YY139 pKa = 10.93EE140 pKa = 4.26LQNGEE145 pKa = 4.91LIVSYY150 pKa = 10.64FDD152 pKa = 3.94EE153 pKa = 4.65NGNRR157 pKa = 11.84QPNAVDD163 pKa = 3.02GGTFRR168 pKa = 11.84TILSYY173 pKa = 11.24DD174 pKa = 3.5EE175 pKa = 5.39SSDD178 pKa = 3.61SFLVADD184 pKa = 4.45YY185 pKa = 8.17FTDD188 pKa = 3.07TRR190 pKa = 11.84TRR192 pKa = 11.84QMDD195 pKa = 3.58PVVIARR201 pKa = 11.84EE202 pKa = 3.9DD203 pKa = 3.49LLRR206 pKa = 11.84WNIEE210 pKa = 3.93NRR212 pKa = 11.84EE213 pKa = 4.01GTYY216 pKa = 10.6VYY218 pKa = 10.9YY219 pKa = 11.12NEE221 pKa = 4.31AQEE224 pKa = 3.83ITAQEE229 pKa = 4.66EE230 pKa = 4.58YY231 pKa = 11.16AQGKK235 pKa = 9.77LNGTVTFWYY244 pKa = 10.32QDD246 pKa = 3.06TGLPAQKK253 pKa = 9.62MEE255 pKa = 4.98EE256 pKa = 4.15INFKK260 pKa = 10.72DD261 pKa = 3.82GQQEE265 pKa = 4.0GLYY268 pKa = 10.31VSWYY272 pKa = 8.59PNGYY276 pKa = 7.27PQYY279 pKa = 10.23RR280 pKa = 11.84INFTDD285 pKa = 3.23MGIDD289 pKa = 5.07SVACWDD295 pKa = 3.7EE296 pKa = 4.15NFVAGTEE303 pKa = 4.14DD304 pKa = 3.13EE305 pKa = 4.49CMARR309 pKa = 11.84LTGAAEE315 pKa = 3.98AAEE318 pKa = 4.41VEE320 pKa = 4.53ANANDD325 pKa = 4.73EE326 pKa = 4.47INDD329 pKa = 4.0DD330 pKa = 4.33LDD332 pKa = 3.88VEE334 pKa = 4.71IIIPSDD340 pKa = 3.44QVVTPTVEE348 pKa = 4.09NQSDD352 pKa = 4.28LEE354 pKa = 4.24DD355 pKa = 5.86AITPDD360 pKa = 3.13ATVLFPHH367 pKa = 7.43DD368 pKa = 3.98EE369 pKa = 4.28DD370 pKa = 4.05NRR372 pKa = 11.84ILIDD376 pKa = 4.28DD377 pKa = 4.3ADD379 pKa = 3.98EE380 pKa = 5.04AFDD383 pKa = 3.96SAVDD387 pKa = 3.76TTEE390 pKa = 3.96TTPALTDD397 pKa = 3.34EE398 pKa = 4.88EE399 pKa = 4.2EE400 pKa = 4.28AADD403 pKa = 3.53IEE405 pKa = 4.5AVEE408 pKa = 4.46EE409 pKa = 4.29ADD411 pKa = 5.34DD412 pKa = 4.06VQIPNQPEE420 pKa = 4.06PEE422 pKa = 4.29TEE424 pKa = 3.98GKK426 pKa = 10.49RR427 pKa = 11.84ILRR430 pKa = 11.84SLNNILEE437 pKa = 4.28ALL439 pKa = 3.9
MM1 pKa = 7.55TYY3 pKa = 10.67KK4 pKa = 9.54KK5 pKa = 8.29TALSIASLLILGTFSYY21 pKa = 10.61AQQTPTEE28 pKa = 4.44LEE30 pKa = 3.97PNGIIAYY37 pKa = 9.15FAEE40 pKa = 5.6DD41 pKa = 3.51GTLLPSRR48 pKa = 11.84EE49 pKa = 3.43GSEE52 pKa = 4.5YY53 pKa = 10.03YY54 pKa = 10.27RR55 pKa = 11.84HH56 pKa = 6.13FKK58 pKa = 10.89DD59 pKa = 4.05VVSGCYY65 pKa = 9.46LVQDD69 pKa = 5.43FYY71 pKa = 11.99SQNDD75 pKa = 3.44QKK77 pKa = 10.24QTDD80 pKa = 5.09PICFMDD86 pKa = 4.54PGEE89 pKa = 4.13LQSWFPQSLYY99 pKa = 11.12GPLTFWHH106 pKa = 6.98KK107 pKa = 11.04DD108 pKa = 3.01GTKK111 pKa = 9.96AQSGYY116 pKa = 9.12SQQDD120 pKa = 3.05GGGNGTWTTWDD131 pKa = 3.23QDD133 pKa = 3.85GNASDD138 pKa = 5.29YY139 pKa = 10.93EE140 pKa = 4.26LQNGEE145 pKa = 4.91LIVSYY150 pKa = 10.64FDD152 pKa = 3.94EE153 pKa = 4.65NGNRR157 pKa = 11.84QPNAVDD163 pKa = 3.02GGTFRR168 pKa = 11.84TILSYY173 pKa = 11.24DD174 pKa = 3.5EE175 pKa = 5.39SSDD178 pKa = 3.61SFLVADD184 pKa = 4.45YY185 pKa = 8.17FTDD188 pKa = 3.07TRR190 pKa = 11.84TRR192 pKa = 11.84QMDD195 pKa = 3.58PVVIARR201 pKa = 11.84EE202 pKa = 3.9DD203 pKa = 3.49LLRR206 pKa = 11.84WNIEE210 pKa = 3.93NRR212 pKa = 11.84EE213 pKa = 4.01GTYY216 pKa = 10.6VYY218 pKa = 10.9YY219 pKa = 11.12NEE221 pKa = 4.31AQEE224 pKa = 3.83ITAQEE229 pKa = 4.66EE230 pKa = 4.58YY231 pKa = 11.16AQGKK235 pKa = 9.77LNGTVTFWYY244 pKa = 10.32QDD246 pKa = 3.06TGLPAQKK253 pKa = 9.62MEE255 pKa = 4.98EE256 pKa = 4.15INFKK260 pKa = 10.72DD261 pKa = 3.82GQQEE265 pKa = 4.0GLYY268 pKa = 10.31VSWYY272 pKa = 8.59PNGYY276 pKa = 7.27PQYY279 pKa = 10.23RR280 pKa = 11.84INFTDD285 pKa = 3.23MGIDD289 pKa = 5.07SVACWDD295 pKa = 3.7EE296 pKa = 4.15NFVAGTEE303 pKa = 4.14DD304 pKa = 3.13EE305 pKa = 4.49CMARR309 pKa = 11.84LTGAAEE315 pKa = 3.98AAEE318 pKa = 4.41VEE320 pKa = 4.53ANANDD325 pKa = 4.73EE326 pKa = 4.47INDD329 pKa = 4.0DD330 pKa = 4.33LDD332 pKa = 3.88VEE334 pKa = 4.71IIIPSDD340 pKa = 3.44QVVTPTVEE348 pKa = 4.09NQSDD352 pKa = 4.28LEE354 pKa = 4.24DD355 pKa = 5.86AITPDD360 pKa = 3.13ATVLFPHH367 pKa = 7.43DD368 pKa = 3.98EE369 pKa = 4.28DD370 pKa = 4.05NRR372 pKa = 11.84ILIDD376 pKa = 4.28DD377 pKa = 4.3ADD379 pKa = 3.98EE380 pKa = 5.04AFDD383 pKa = 3.96SAVDD387 pKa = 3.76TTEE390 pKa = 3.96TTPALTDD397 pKa = 3.34EE398 pKa = 4.88EE399 pKa = 4.2EE400 pKa = 4.28AADD403 pKa = 3.53IEE405 pKa = 4.5AVEE408 pKa = 4.46EE409 pKa = 4.29ADD411 pKa = 5.34DD412 pKa = 4.06VQIPNQPEE420 pKa = 4.06PEE422 pKa = 4.29TEE424 pKa = 3.98GKK426 pKa = 10.49RR427 pKa = 11.84ILRR430 pKa = 11.84SLNNILEE437 pKa = 4.28ALL439 pKa = 3.9
Molecular weight: 49.34 kDa
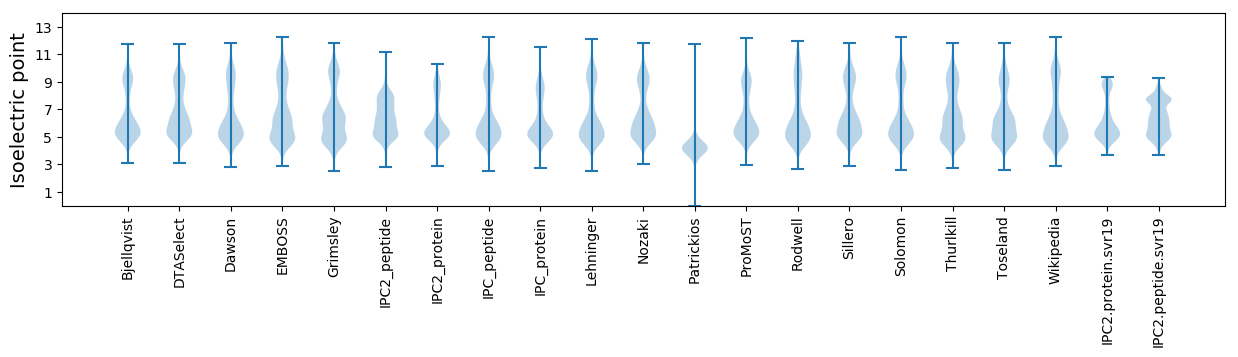
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L8XXF5|L8XXF5_9GAMM Cyclopropane-fatty-acyl-phospholipid synthase OS=Wohlfahrtiimonas chitiniclastica SH04 OX=1261130 GN=F387_01198 PE=3 SV=1
MM1 pKa = 7.69AVNQNYY7 pKa = 7.53GTGRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.42TSAARR18 pKa = 11.84VFLRR22 pKa = 11.84KK23 pKa = 8.53GTGNIVVNGKK33 pKa = 8.73PLDD36 pKa = 3.66EE37 pKa = 4.25YY38 pKa = 10.96FGRR41 pKa = 11.84KK42 pKa = 5.21TACMVVRR49 pKa = 11.84QPLEE53 pKa = 3.91LLDD56 pKa = 3.72ATEE59 pKa = 4.01QFDD62 pKa = 4.17VYY64 pKa = 9.71VTVVGGGQTGQSGAIRR80 pKa = 11.84HH81 pKa = 5.65GLTRR85 pKa = 11.84ALIEE89 pKa = 3.9YY90 pKa = 10.65DD91 pKa = 3.51EE92 pKa = 4.72VNRR95 pKa = 11.84SALRR99 pKa = 11.84KK100 pKa = 9.23AGYY103 pKa = 6.29VTRR106 pKa = 11.84DD107 pKa = 3.04ARR109 pKa = 11.84EE110 pKa = 3.89VEE112 pKa = 4.21RR113 pKa = 11.84KK114 pKa = 9.64KK115 pKa = 11.05VGLRR119 pKa = 11.84KK120 pKa = 9.46ARR122 pKa = 11.84RR123 pKa = 11.84ATQFSKK129 pKa = 10.98RR130 pKa = 3.61
MM1 pKa = 7.69AVNQNYY7 pKa = 7.53GTGRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.42TSAARR18 pKa = 11.84VFLRR22 pKa = 11.84KK23 pKa = 8.53GTGNIVVNGKK33 pKa = 8.73PLDD36 pKa = 3.66EE37 pKa = 4.25YY38 pKa = 10.96FGRR41 pKa = 11.84KK42 pKa = 5.21TACMVVRR49 pKa = 11.84QPLEE53 pKa = 3.91LLDD56 pKa = 3.72ATEE59 pKa = 4.01QFDD62 pKa = 4.17VYY64 pKa = 9.71VTVVGGGQTGQSGAIRR80 pKa = 11.84HH81 pKa = 5.65GLTRR85 pKa = 11.84ALIEE89 pKa = 3.9YY90 pKa = 10.65DD91 pKa = 3.51EE92 pKa = 4.72VNRR95 pKa = 11.84SALRR99 pKa = 11.84KK100 pKa = 9.23AGYY103 pKa = 6.29VTRR106 pKa = 11.84DD107 pKa = 3.04ARR109 pKa = 11.84EE110 pKa = 3.89VEE112 pKa = 4.21RR113 pKa = 11.84KK114 pKa = 9.64KK115 pKa = 11.05VGLRR119 pKa = 11.84KK120 pKa = 9.46ARR122 pKa = 11.84RR123 pKa = 11.84ATQFSKK129 pKa = 10.98RR130 pKa = 3.61
Molecular weight: 14.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
650783 |
14 |
4331 |
324.7 |
36.12 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.881 ± 0.061 | 0.893 ± 0.018 |
5.504 ± 0.05 | 6.08 ± 0.069 |
4.228 ± 0.041 | 6.735 ± 0.09 |
2.494 ± 0.028 | 7.312 ± 0.05 |
5.516 ± 0.046 | 10.33 ± 0.065 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.826 ± 0.031 | 4.364 ± 0.064 |
3.934 ± 0.041 | 4.085 ± 0.041 |
4.341 ± 0.048 | 5.771 ± 0.038 |
5.667 ± 0.063 | 6.738 ± 0.048 |
1.086 ± 0.02 | 3.215 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
