
Maize Iranian mosaic nucleorhabdovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Nucleorhabdovirus
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
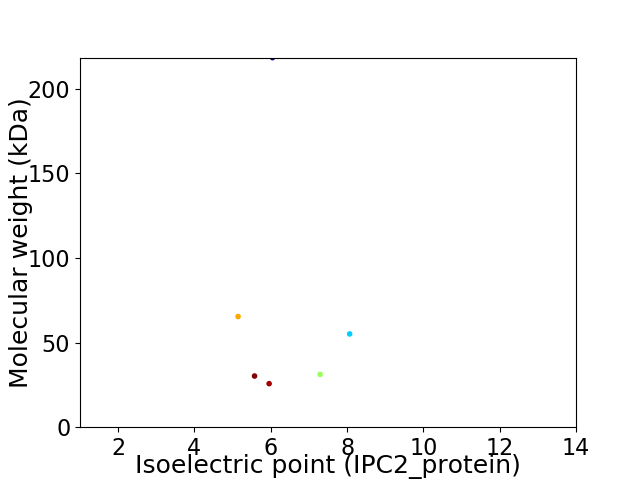
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|B5FX46|B5FX46_9RHAB Large structural protein OS=Maize Iranian mosaic nucleorhabdovirus OX=348823 GN=L PE=4 SV=1
MM1 pKa = 7.19ATSFLLLLSLSSILAISATSAPALTSGRR29 pKa = 11.84IPRR32 pKa = 11.84DD33 pKa = 3.48EE34 pKa = 4.82EE35 pKa = 4.03PSSDD39 pKa = 4.43KK40 pKa = 11.14GDD42 pKa = 3.45SDD44 pKa = 5.46LYY46 pKa = 10.56PLYY49 pKa = 10.72EE50 pKa = 4.59CGKK53 pKa = 9.3EE54 pKa = 4.22GTGVPITSWYY64 pKa = 8.14GACRR68 pKa = 11.84GACSMASNSTEE79 pKa = 3.97VVAEE83 pKa = 4.01IYY85 pKa = 10.23YY86 pKa = 10.79RR87 pKa = 11.84NDD89 pKa = 2.83SVGYY93 pKa = 9.8IDD95 pKa = 4.01VLSVRR100 pKa = 11.84TTEE103 pKa = 3.87IKK105 pKa = 9.38KK106 pKa = 8.25TSHH109 pKa = 4.34VTWYY113 pKa = 9.65GACDD117 pKa = 3.38EE118 pKa = 4.3EE119 pKa = 5.06TIVGANSLAPEE130 pKa = 4.21YY131 pKa = 10.32IIEE134 pKa = 3.92QSIEE138 pKa = 3.99FLHH141 pKa = 6.8SGLDD145 pKa = 3.29SWQYY149 pKa = 9.94GADD152 pKa = 3.06IHH154 pKa = 6.42VHH156 pKa = 5.18DD157 pKa = 5.45TIGFPDD163 pKa = 4.86CDD165 pKa = 3.9YY166 pKa = 11.75LDD168 pKa = 4.58DD169 pKa = 4.97KK170 pKa = 11.82NNEE173 pKa = 3.33GWRR176 pKa = 11.84FIITKK181 pKa = 10.08RR182 pKa = 11.84AIEE185 pKa = 4.38LKK187 pKa = 10.54SDD189 pKa = 3.2IAGEE193 pKa = 4.45GYY195 pKa = 10.04IVDD198 pKa = 4.36PDD200 pKa = 3.69LGYY203 pKa = 11.12VFPVSDD209 pKa = 4.55GIGRR213 pKa = 11.84GKK215 pKa = 10.16FWYY218 pKa = 8.42IWKK221 pKa = 10.16SSSVPSGGCFFKK233 pKa = 11.06SAGTGNCTLLMDD245 pKa = 4.65TFTYY249 pKa = 10.26SCPEE253 pKa = 3.97LNVAFSAKK261 pKa = 10.31VNTKK265 pKa = 10.83LEE267 pKa = 4.41GTCVGDD273 pKa = 4.8LNISSDD279 pKa = 3.63GVSYY283 pKa = 11.17ALRR286 pKa = 11.84SQQSTGSVSSALINLWHH303 pKa = 6.25QSQEE307 pKa = 3.86AVTQQLILVINDD319 pKa = 3.54ALGKK323 pKa = 10.03IEE325 pKa = 4.86SSYY328 pKa = 11.64CEE330 pKa = 3.95ASCDD334 pKa = 3.39LTEE337 pKa = 4.0ALFYY341 pKa = 10.77RR342 pKa = 11.84VTDD345 pKa = 3.64NPAVVEE351 pKa = 4.51TPVGPWLPSLSKK363 pKa = 10.7GKK365 pKa = 10.71VSIIPCQSEE374 pKa = 3.85QQLIIILPIEE384 pKa = 3.95ICYY387 pKa = 10.18EE388 pKa = 3.76PFMLKK393 pKa = 10.11VKK395 pKa = 9.82SLKK398 pKa = 9.84TGEE401 pKa = 4.41TFWWLPSHH409 pKa = 5.91SHH411 pKa = 4.64VTSSTSCRR419 pKa = 11.84SIATEE424 pKa = 3.59EE425 pKa = 4.4VEE427 pKa = 4.95FQGEE431 pKa = 4.33LKK433 pKa = 10.75KK434 pKa = 10.61PLTFEE439 pKa = 4.61FWRR442 pKa = 11.84GMYY445 pKa = 10.29LLPYY449 pKa = 9.05PYY451 pKa = 9.65NGSGTWIDD459 pKa = 3.49NPGTHH464 pKa = 6.11IHH466 pKa = 7.04RR467 pKa = 11.84SSKK470 pKa = 9.23WFPSINQLSYY480 pKa = 11.1SVPIKK485 pKa = 10.75LPLITKK491 pKa = 7.95EE492 pKa = 3.78IRR494 pKa = 11.84KK495 pKa = 8.68HH496 pKa = 4.24VQQTVVSIGGFSNITSHH513 pKa = 7.74PITEE517 pKa = 5.15GIWTALSTAGLIIGKK532 pKa = 7.41TAARR536 pKa = 11.84VVMWWSEE543 pKa = 3.92LEE545 pKa = 4.23EE546 pKa = 4.25TVKK549 pKa = 11.04GYY551 pKa = 8.25VTLAGGVIISLLVGYY566 pKa = 10.71LMLKK570 pKa = 9.79TILLTRR576 pKa = 11.84RR577 pKa = 11.84PSYY580 pKa = 10.16SAVPRR585 pKa = 11.84EE586 pKa = 4.41VNWITPAKK594 pKa = 10.22
MM1 pKa = 7.19ATSFLLLLSLSSILAISATSAPALTSGRR29 pKa = 11.84IPRR32 pKa = 11.84DD33 pKa = 3.48EE34 pKa = 4.82EE35 pKa = 4.03PSSDD39 pKa = 4.43KK40 pKa = 11.14GDD42 pKa = 3.45SDD44 pKa = 5.46LYY46 pKa = 10.56PLYY49 pKa = 10.72EE50 pKa = 4.59CGKK53 pKa = 9.3EE54 pKa = 4.22GTGVPITSWYY64 pKa = 8.14GACRR68 pKa = 11.84GACSMASNSTEE79 pKa = 3.97VVAEE83 pKa = 4.01IYY85 pKa = 10.23YY86 pKa = 10.79RR87 pKa = 11.84NDD89 pKa = 2.83SVGYY93 pKa = 9.8IDD95 pKa = 4.01VLSVRR100 pKa = 11.84TTEE103 pKa = 3.87IKK105 pKa = 9.38KK106 pKa = 8.25TSHH109 pKa = 4.34VTWYY113 pKa = 9.65GACDD117 pKa = 3.38EE118 pKa = 4.3EE119 pKa = 5.06TIVGANSLAPEE130 pKa = 4.21YY131 pKa = 10.32IIEE134 pKa = 3.92QSIEE138 pKa = 3.99FLHH141 pKa = 6.8SGLDD145 pKa = 3.29SWQYY149 pKa = 9.94GADD152 pKa = 3.06IHH154 pKa = 6.42VHH156 pKa = 5.18DD157 pKa = 5.45TIGFPDD163 pKa = 4.86CDD165 pKa = 3.9YY166 pKa = 11.75LDD168 pKa = 4.58DD169 pKa = 4.97KK170 pKa = 11.82NNEE173 pKa = 3.33GWRR176 pKa = 11.84FIITKK181 pKa = 10.08RR182 pKa = 11.84AIEE185 pKa = 4.38LKK187 pKa = 10.54SDD189 pKa = 3.2IAGEE193 pKa = 4.45GYY195 pKa = 10.04IVDD198 pKa = 4.36PDD200 pKa = 3.69LGYY203 pKa = 11.12VFPVSDD209 pKa = 4.55GIGRR213 pKa = 11.84GKK215 pKa = 10.16FWYY218 pKa = 8.42IWKK221 pKa = 10.16SSSVPSGGCFFKK233 pKa = 11.06SAGTGNCTLLMDD245 pKa = 4.65TFTYY249 pKa = 10.26SCPEE253 pKa = 3.97LNVAFSAKK261 pKa = 10.31VNTKK265 pKa = 10.83LEE267 pKa = 4.41GTCVGDD273 pKa = 4.8LNISSDD279 pKa = 3.63GVSYY283 pKa = 11.17ALRR286 pKa = 11.84SQQSTGSVSSALINLWHH303 pKa = 6.25QSQEE307 pKa = 3.86AVTQQLILVINDD319 pKa = 3.54ALGKK323 pKa = 10.03IEE325 pKa = 4.86SSYY328 pKa = 11.64CEE330 pKa = 3.95ASCDD334 pKa = 3.39LTEE337 pKa = 4.0ALFYY341 pKa = 10.77RR342 pKa = 11.84VTDD345 pKa = 3.64NPAVVEE351 pKa = 4.51TPVGPWLPSLSKK363 pKa = 10.7GKK365 pKa = 10.71VSIIPCQSEE374 pKa = 3.85QQLIIILPIEE384 pKa = 3.95ICYY387 pKa = 10.18EE388 pKa = 3.76PFMLKK393 pKa = 10.11VKK395 pKa = 9.82SLKK398 pKa = 9.84TGEE401 pKa = 4.41TFWWLPSHH409 pKa = 5.91SHH411 pKa = 4.64VTSSTSCRR419 pKa = 11.84SIATEE424 pKa = 3.59EE425 pKa = 4.4VEE427 pKa = 4.95FQGEE431 pKa = 4.33LKK433 pKa = 10.75KK434 pKa = 10.61PLTFEE439 pKa = 4.61FWRR442 pKa = 11.84GMYY445 pKa = 10.29LLPYY449 pKa = 9.05PYY451 pKa = 9.65NGSGTWIDD459 pKa = 3.49NPGTHH464 pKa = 6.11IHH466 pKa = 7.04RR467 pKa = 11.84SSKK470 pKa = 9.23WFPSINQLSYY480 pKa = 11.1SVPIKK485 pKa = 10.75LPLITKK491 pKa = 7.95EE492 pKa = 3.78IRR494 pKa = 11.84KK495 pKa = 8.68HH496 pKa = 4.24VQQTVVSIGGFSNITSHH513 pKa = 7.74PITEE517 pKa = 5.15GIWTALSTAGLIIGKK532 pKa = 7.41TAARR536 pKa = 11.84VVMWWSEE543 pKa = 3.92LEE545 pKa = 4.23EE546 pKa = 4.25TVKK549 pKa = 11.04GYY551 pKa = 8.25VTLAGGVIISLLVGYY566 pKa = 10.71LMLKK570 pKa = 9.79TILLTRR576 pKa = 11.84RR577 pKa = 11.84PSYY580 pKa = 10.16SAVPRR585 pKa = 11.84EE586 pKa = 4.41VNWITPAKK594 pKa = 10.22
Molecular weight: 65.37 kDa
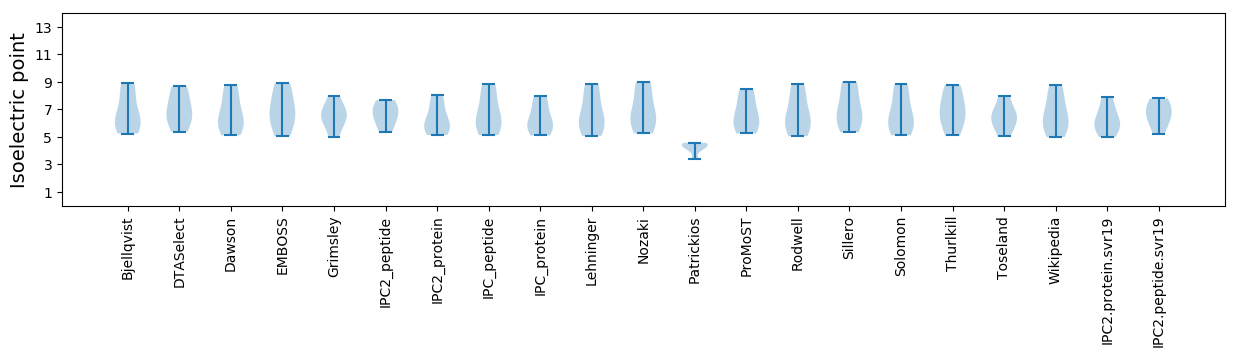
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B5FX42|B5FX42_9RHAB Phosphoprotein OS=Maize Iranian mosaic nucleorhabdovirus OX=348823 GN=P PE=4 SV=1
MM1 pKa = 7.65ASITIPVEE9 pKa = 3.91QVTKK13 pKa = 10.7FSDD16 pKa = 3.71DD17 pKa = 3.69VKK19 pKa = 11.44SLTQKK24 pKa = 10.69ASEE27 pKa = 4.25IPSSKK32 pKa = 10.64SIIPQTEE39 pKa = 3.78FSDD42 pKa = 4.63AEE44 pKa = 4.13LKK46 pKa = 9.29KK47 pKa = 9.02TLKK50 pKa = 9.79FWEE53 pKa = 4.48VTAMDD58 pKa = 5.04DD59 pKa = 3.87ATMVTQWSGVQSALAGGTFSAQNLKK84 pKa = 9.77EE85 pKa = 3.98LCQLAFNLRR94 pKa = 11.84KK95 pKa = 8.87PHH97 pKa = 6.15EE98 pKa = 4.77PGHH101 pKa = 5.53VFIHH105 pKa = 6.76NIPDD109 pKa = 2.81SWKK112 pKa = 10.42AYY114 pKa = 10.62LEE116 pKa = 4.19AATPDD121 pKa = 3.46GTAIPATEE129 pKa = 4.38NEE131 pKa = 4.52STMSTIGAAAATGSADD147 pKa = 3.07EE148 pKa = 4.21TAPNKK153 pKa = 9.95AAAIAFLSCTLMRR166 pKa = 11.84LAIKK170 pKa = 9.68EE171 pKa = 4.39PKK173 pKa = 10.03HH174 pKa = 8.42IMDD177 pKa = 4.06AMTNIRR183 pKa = 11.84SRR185 pKa = 11.84YY186 pKa = 7.21GSLYY190 pKa = 9.97GRR192 pKa = 11.84SSPFLNSVTFNLSQLSRR209 pKa = 11.84IKK211 pKa = 10.57QGLEE215 pKa = 3.73TYY217 pKa = 10.45AVARR221 pKa = 11.84GTLFYY226 pKa = 9.17YY227 pKa = 9.91TRR229 pKa = 11.84HH230 pKa = 6.24ADD232 pKa = 3.05STYY235 pKa = 9.93TYY237 pKa = 7.52TTPSYY242 pKa = 10.34GICRR246 pKa = 11.84YY247 pKa = 10.24LLFQHH252 pKa = 6.92LEE254 pKa = 4.07LEE256 pKa = 4.52GMHH259 pKa = 6.77IYY261 pKa = 11.03KK262 pKa = 9.46MLLALQTEE270 pKa = 4.93WATVPMRR277 pKa = 11.84LLLTWIRR284 pKa = 11.84NPRR287 pKa = 11.84SKK289 pKa = 10.7LAVLEE294 pKa = 3.97IVKK297 pKa = 10.25ILTNWDD303 pKa = 3.3KK304 pKa = 12.01ANVDD308 pKa = 4.02KK309 pKa = 10.95GWKK312 pKa = 8.36YY313 pKa = 11.18ARR315 pKa = 11.84LVNNTFFLNLSSRR328 pKa = 11.84RR329 pKa = 11.84NTYY332 pKa = 10.31LCAVLAALNKK342 pKa = 10.06KK343 pKa = 9.01FVPQGTGDD351 pKa = 3.54YY352 pKa = 11.05ANPNNIAVIKK362 pKa = 11.03NMDD365 pKa = 3.86SAVKK369 pKa = 10.1KK370 pKa = 10.17QVSPDD375 pKa = 3.37VQIVEE380 pKa = 4.8RR381 pKa = 11.84IYY383 pKa = 10.78DD384 pKa = 3.45IFLASSGSDD393 pKa = 3.3DD394 pKa = 4.21AGTAFTLSRR403 pKa = 11.84GVKK406 pKa = 8.58RR407 pKa = 11.84TSPNNPEE414 pKa = 4.24TRR416 pKa = 11.84MQWTPTQEE424 pKa = 4.12LEE426 pKa = 4.49RR427 pKa = 11.84DD428 pKa = 3.59LEE430 pKa = 4.25NPRR433 pKa = 11.84LRR435 pKa = 11.84RR436 pKa = 11.84GSRR439 pKa = 11.84IRR441 pKa = 11.84EE442 pKa = 4.09VNCRR446 pKa = 11.84PEE448 pKa = 4.5SDD450 pKa = 3.09MSQMQRR456 pKa = 11.84EE457 pKa = 4.57LVFHH461 pKa = 6.58EE462 pKa = 4.57SVSVSILCDD471 pKa = 3.3EE472 pKa = 4.59GRR474 pKa = 11.84VMSDD478 pKa = 3.7PITRR482 pKa = 11.84VSSIKK487 pKa = 10.75CGVLLLL493 pKa = 4.39
MM1 pKa = 7.65ASITIPVEE9 pKa = 3.91QVTKK13 pKa = 10.7FSDD16 pKa = 3.71DD17 pKa = 3.69VKK19 pKa = 11.44SLTQKK24 pKa = 10.69ASEE27 pKa = 4.25IPSSKK32 pKa = 10.64SIIPQTEE39 pKa = 3.78FSDD42 pKa = 4.63AEE44 pKa = 4.13LKK46 pKa = 9.29KK47 pKa = 9.02TLKK50 pKa = 9.79FWEE53 pKa = 4.48VTAMDD58 pKa = 5.04DD59 pKa = 3.87ATMVTQWSGVQSALAGGTFSAQNLKK84 pKa = 9.77EE85 pKa = 3.98LCQLAFNLRR94 pKa = 11.84KK95 pKa = 8.87PHH97 pKa = 6.15EE98 pKa = 4.77PGHH101 pKa = 5.53VFIHH105 pKa = 6.76NIPDD109 pKa = 2.81SWKK112 pKa = 10.42AYY114 pKa = 10.62LEE116 pKa = 4.19AATPDD121 pKa = 3.46GTAIPATEE129 pKa = 4.38NEE131 pKa = 4.52STMSTIGAAAATGSADD147 pKa = 3.07EE148 pKa = 4.21TAPNKK153 pKa = 9.95AAAIAFLSCTLMRR166 pKa = 11.84LAIKK170 pKa = 9.68EE171 pKa = 4.39PKK173 pKa = 10.03HH174 pKa = 8.42IMDD177 pKa = 4.06AMTNIRR183 pKa = 11.84SRR185 pKa = 11.84YY186 pKa = 7.21GSLYY190 pKa = 9.97GRR192 pKa = 11.84SSPFLNSVTFNLSQLSRR209 pKa = 11.84IKK211 pKa = 10.57QGLEE215 pKa = 3.73TYY217 pKa = 10.45AVARR221 pKa = 11.84GTLFYY226 pKa = 9.17YY227 pKa = 9.91TRR229 pKa = 11.84HH230 pKa = 6.24ADD232 pKa = 3.05STYY235 pKa = 9.93TYY237 pKa = 7.52TTPSYY242 pKa = 10.34GICRR246 pKa = 11.84YY247 pKa = 10.24LLFQHH252 pKa = 6.92LEE254 pKa = 4.07LEE256 pKa = 4.52GMHH259 pKa = 6.77IYY261 pKa = 11.03KK262 pKa = 9.46MLLALQTEE270 pKa = 4.93WATVPMRR277 pKa = 11.84LLLTWIRR284 pKa = 11.84NPRR287 pKa = 11.84SKK289 pKa = 10.7LAVLEE294 pKa = 3.97IVKK297 pKa = 10.25ILTNWDD303 pKa = 3.3KK304 pKa = 12.01ANVDD308 pKa = 4.02KK309 pKa = 10.95GWKK312 pKa = 8.36YY313 pKa = 11.18ARR315 pKa = 11.84LVNNTFFLNLSSRR328 pKa = 11.84RR329 pKa = 11.84NTYY332 pKa = 10.31LCAVLAALNKK342 pKa = 10.06KK343 pKa = 9.01FVPQGTGDD351 pKa = 3.54YY352 pKa = 11.05ANPNNIAVIKK362 pKa = 11.03NMDD365 pKa = 3.86SAVKK369 pKa = 10.1KK370 pKa = 10.17QVSPDD375 pKa = 3.37VQIVEE380 pKa = 4.8RR381 pKa = 11.84IYY383 pKa = 10.78DD384 pKa = 3.45IFLASSGSDD393 pKa = 3.3DD394 pKa = 4.21AGTAFTLSRR403 pKa = 11.84GVKK406 pKa = 8.58RR407 pKa = 11.84TSPNNPEE414 pKa = 4.24TRR416 pKa = 11.84MQWTPTQEE424 pKa = 4.12LEE426 pKa = 4.49RR427 pKa = 11.84DD428 pKa = 3.59LEE430 pKa = 4.25NPRR433 pKa = 11.84LRR435 pKa = 11.84RR436 pKa = 11.84GSRR439 pKa = 11.84IRR441 pKa = 11.84EE442 pKa = 4.09VNCRR446 pKa = 11.84PEE448 pKa = 4.5SDD450 pKa = 3.09MSQMQRR456 pKa = 11.84EE457 pKa = 4.57LVFHH461 pKa = 6.58EE462 pKa = 4.57SVSVSILCDD471 pKa = 3.3EE472 pKa = 4.59GRR474 pKa = 11.84VMSDD478 pKa = 3.7PITRR482 pKa = 11.84VSSIKK487 pKa = 10.75CGVLLLL493 pKa = 4.39
Molecular weight: 55.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3798 |
233 |
1926 |
633.0 |
70.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.319 ± 0.634 | 1.606 ± 0.201 |
5.608 ± 0.56 | 6.135 ± 0.203 |
3.107 ± 0.264 | 6.214 ± 0.454 |
2.396 ± 0.268 | 7.399 ± 0.467 |
5.292 ± 0.237 | 9.9 ± 0.468 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.975 ± 0.449 | 3.923 ± 0.344 |
4.371 ± 0.469 | 2.87 ± 0.249 |
5.687 ± 0.69 | 8.557 ± 0.735 |
6.477 ± 0.756 | 6.214 ± 0.282 |
1.711 ± 0.283 | 3.239 ± 0.245 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
