
Legionella quinlivanii
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Legionellales; Legionellaceae; Legionella
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
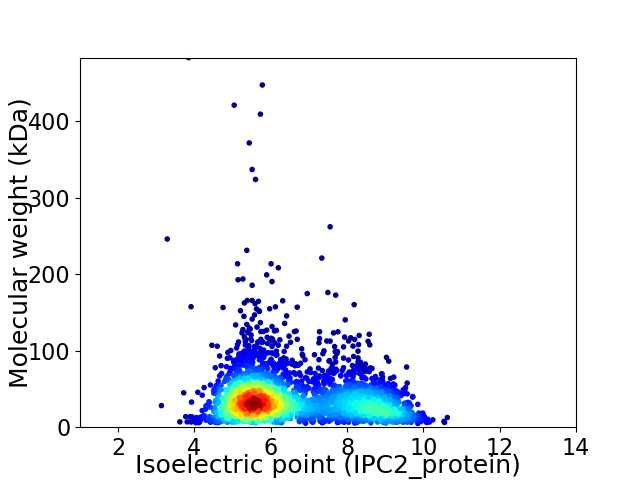
Virtual 2D-PAGE plot for 2989 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0W0XY50|A0A0W0XY50_9GAMM Hemolysin D OS=Legionella quinlivanii OX=45073 GN=Lqui_1914 PE=4 SV=1
MM1 pKa = 7.35AVAVNSGIFSQLALNTQFEE20 pKa = 4.77LSQVTRR26 pKa = 11.84ILPLFLSNTLVVSINNSFFSVQRR49 pKa = 11.84PVYY52 pKa = 9.46PPADD56 pKa = 3.23VAEE59 pKa = 4.31RR60 pKa = 11.84DD61 pKa = 3.79EE62 pKa = 4.53GVKK65 pKa = 10.22QADD68 pKa = 3.4LDD70 pKa = 4.1LFNYY74 pKa = 9.91LLNFLPPSYY83 pKa = 10.65LIVEE87 pKa = 4.63PDD89 pKa = 3.46NLDD92 pKa = 3.3GNTFVSFNPYY102 pKa = 9.94YY103 pKa = 10.78SLGYY107 pKa = 10.14VDD109 pKa = 5.63AGYY112 pKa = 7.94DD113 pKa = 3.33TLGISQSIINTLLDD127 pKa = 2.98NWFFLSRR134 pKa = 11.84RR135 pKa = 11.84LTLLNSDD142 pKa = 3.41ADD144 pKa = 4.23EE145 pKa = 4.71NNSEE149 pKa = 4.14SSNEE153 pKa = 3.81SRR155 pKa = 11.84EE156 pKa = 4.25SNASVNVSLFEE167 pKa = 4.66SGLQNGHH174 pKa = 6.78APGAATAYY182 pKa = 10.31SGNLSLLNPEE192 pKa = 4.39NSPVALAAGSGVLPFTSNDD211 pKa = 2.69IPINYY216 pKa = 9.09IIEE219 pKa = 4.21NSQLIAQAGGEE230 pKa = 4.52TIFILQILPNNQFSFQLNNNLDD252 pKa = 3.82HH253 pKa = 7.32PLPGADD259 pKa = 4.18QIVIDD264 pKa = 4.76LSEE267 pKa = 5.11FIQIGNINLPPGTFIIRR284 pKa = 11.84INDD287 pKa = 4.68DD288 pKa = 3.01IPLAVAGSNQQSLLHH303 pKa = 6.62EE304 pKa = 4.82DD305 pKa = 4.06ALPGANIDD313 pKa = 3.89TPNPLSEE320 pKa = 3.98ISGALSALVSFGADD334 pKa = 3.19GQAGFHH340 pKa = 6.98FISEE344 pKa = 4.34PAISTVLEE352 pKa = 4.23SFGLSSGGQPLDD364 pKa = 3.42YY365 pKa = 10.56QVADD369 pKa = 3.72NRR371 pKa = 11.84LQGLDD376 pKa = 3.42PDD378 pKa = 4.25GNEE381 pKa = 3.77VFTLIVNTDD390 pKa = 2.73GSYY393 pKa = 10.66QFILQQSLDD402 pKa = 3.89HH403 pKa = 6.66LGSNASTILFPWVFF417 pKa = 3.42
MM1 pKa = 7.35AVAVNSGIFSQLALNTQFEE20 pKa = 4.77LSQVTRR26 pKa = 11.84ILPLFLSNTLVVSINNSFFSVQRR49 pKa = 11.84PVYY52 pKa = 9.46PPADD56 pKa = 3.23VAEE59 pKa = 4.31RR60 pKa = 11.84DD61 pKa = 3.79EE62 pKa = 4.53GVKK65 pKa = 10.22QADD68 pKa = 3.4LDD70 pKa = 4.1LFNYY74 pKa = 9.91LLNFLPPSYY83 pKa = 10.65LIVEE87 pKa = 4.63PDD89 pKa = 3.46NLDD92 pKa = 3.3GNTFVSFNPYY102 pKa = 9.94YY103 pKa = 10.78SLGYY107 pKa = 10.14VDD109 pKa = 5.63AGYY112 pKa = 7.94DD113 pKa = 3.33TLGISQSIINTLLDD127 pKa = 2.98NWFFLSRR134 pKa = 11.84RR135 pKa = 11.84LTLLNSDD142 pKa = 3.41ADD144 pKa = 4.23EE145 pKa = 4.71NNSEE149 pKa = 4.14SSNEE153 pKa = 3.81SRR155 pKa = 11.84EE156 pKa = 4.25SNASVNVSLFEE167 pKa = 4.66SGLQNGHH174 pKa = 6.78APGAATAYY182 pKa = 10.31SGNLSLLNPEE192 pKa = 4.39NSPVALAAGSGVLPFTSNDD211 pKa = 2.69IPINYY216 pKa = 9.09IIEE219 pKa = 4.21NSQLIAQAGGEE230 pKa = 4.52TIFILQILPNNQFSFQLNNNLDD252 pKa = 3.82HH253 pKa = 7.32PLPGADD259 pKa = 4.18QIVIDD264 pKa = 4.76LSEE267 pKa = 5.11FIQIGNINLPPGTFIIRR284 pKa = 11.84INDD287 pKa = 4.68DD288 pKa = 3.01IPLAVAGSNQQSLLHH303 pKa = 6.62EE304 pKa = 4.82DD305 pKa = 4.06ALPGANIDD313 pKa = 3.89TPNPLSEE320 pKa = 3.98ISGALSALVSFGADD334 pKa = 3.19GQAGFHH340 pKa = 6.98FISEE344 pKa = 4.34PAISTVLEE352 pKa = 4.23SFGLSSGGQPLDD364 pKa = 3.42YY365 pKa = 10.56QVADD369 pKa = 3.72NRR371 pKa = 11.84LQGLDD376 pKa = 3.42PDD378 pKa = 4.25GNEE381 pKa = 3.77VFTLIVNTDD390 pKa = 2.73GSYY393 pKa = 10.66QFILQQSLDD402 pKa = 3.89HH403 pKa = 6.66LGSNASTILFPWVFF417 pKa = 3.42
Molecular weight: 44.94 kDa
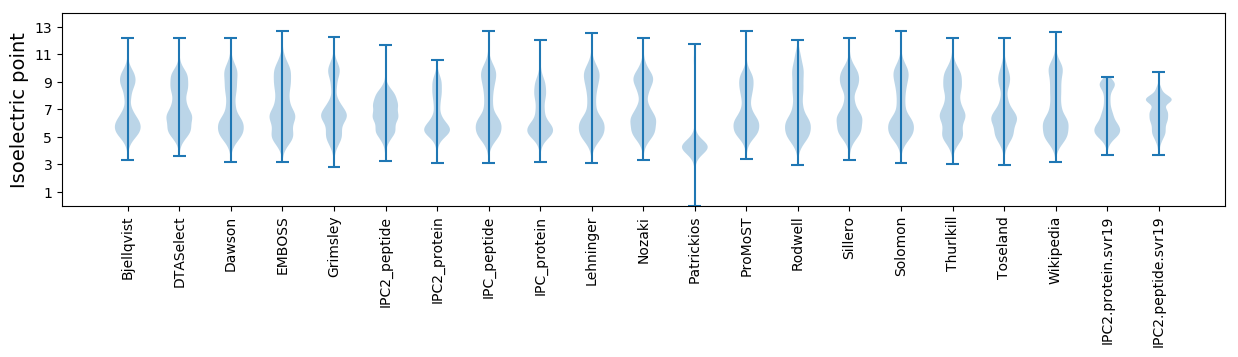
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0W0Y1C9|A0A0W0Y1C9_9GAMM Phosphopentomutase OS=Legionella quinlivanii OX=45073 GN=deoB PE=3 SV=1
MM1 pKa = 7.63ISRR4 pKa = 11.84SVYY7 pKa = 9.23HH8 pKa = 5.04YY9 pKa = 10.22RR10 pKa = 11.84SRR12 pKa = 11.84RR13 pKa = 11.84GTDD16 pKa = 2.52AVVRR20 pKa = 11.84HH21 pKa = 6.87RR22 pKa = 11.84IRR24 pKa = 11.84EE25 pKa = 3.85IAEE28 pKa = 3.67TRR30 pKa = 11.84VRR32 pKa = 11.84YY33 pKa = 9.44GYY35 pKa = 10.32QRR37 pKa = 11.84IHH39 pKa = 6.68VLLQRR44 pKa = 11.84EE45 pKa = 4.41GWIINHH51 pKa = 6.28KK52 pKa = 9.3KK53 pKa = 7.58VHH55 pKa = 6.85RR56 pKa = 11.84IYY58 pKa = 11.03CEE60 pKa = 3.35EE61 pKa = 3.94GLNLRR66 pKa = 11.84RR67 pKa = 11.84KK68 pKa = 9.2RR69 pKa = 11.84PRR71 pKa = 11.84RR72 pKa = 11.84NVAGAHH78 pKa = 5.91RR79 pKa = 11.84EE80 pKa = 4.02DD81 pKa = 4.36RR82 pKa = 11.84SLASALDD89 pKa = 4.08EE90 pKa = 4.4CWSMDD95 pKa = 3.66FVSDD99 pKa = 3.62SLFNGRR105 pKa = 11.84RR106 pKa = 11.84IRR108 pKa = 11.84ALTIVDD114 pKa = 3.57NFSRR118 pKa = 11.84EE119 pKa = 3.93CLAIHH124 pKa = 6.5VGQAIRR130 pKa = 11.84GEE132 pKa = 4.12DD133 pKa = 3.59VACVLEE139 pKa = 4.23GLRR142 pKa = 11.84VMEE145 pKa = 5.03NSGQKK150 pKa = 9.98AA151 pKa = 3.05
MM1 pKa = 7.63ISRR4 pKa = 11.84SVYY7 pKa = 9.23HH8 pKa = 5.04YY9 pKa = 10.22RR10 pKa = 11.84SRR12 pKa = 11.84RR13 pKa = 11.84GTDD16 pKa = 2.52AVVRR20 pKa = 11.84HH21 pKa = 6.87RR22 pKa = 11.84IRR24 pKa = 11.84EE25 pKa = 3.85IAEE28 pKa = 3.67TRR30 pKa = 11.84VRR32 pKa = 11.84YY33 pKa = 9.44GYY35 pKa = 10.32QRR37 pKa = 11.84IHH39 pKa = 6.68VLLQRR44 pKa = 11.84EE45 pKa = 4.41GWIINHH51 pKa = 6.28KK52 pKa = 9.3KK53 pKa = 7.58VHH55 pKa = 6.85RR56 pKa = 11.84IYY58 pKa = 11.03CEE60 pKa = 3.35EE61 pKa = 3.94GLNLRR66 pKa = 11.84RR67 pKa = 11.84KK68 pKa = 9.2RR69 pKa = 11.84PRR71 pKa = 11.84RR72 pKa = 11.84NVAGAHH78 pKa = 5.91RR79 pKa = 11.84EE80 pKa = 4.02DD81 pKa = 4.36RR82 pKa = 11.84SLASALDD89 pKa = 4.08EE90 pKa = 4.4CWSMDD95 pKa = 3.66FVSDD99 pKa = 3.62SLFNGRR105 pKa = 11.84RR106 pKa = 11.84IRR108 pKa = 11.84ALTIVDD114 pKa = 3.57NFSRR118 pKa = 11.84EE119 pKa = 3.93CLAIHH124 pKa = 6.5VGQAIRR130 pKa = 11.84GEE132 pKa = 4.12DD133 pKa = 3.59VACVLEE139 pKa = 4.23GLRR142 pKa = 11.84VMEE145 pKa = 5.03NSGQKK150 pKa = 9.98AA151 pKa = 3.05
Molecular weight: 17.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1064207 |
50 |
4606 |
356.0 |
39.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.982 ± 0.049 | 1.173 ± 0.019 |
4.901 ± 0.03 | 6.247 ± 0.043 |
4.448 ± 0.035 | 6.058 ± 0.047 |
2.375 ± 0.021 | 7.018 ± 0.037 |
5.798 ± 0.043 | 11.193 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.309 ± 0.021 | 4.826 ± 0.038 |
4.171 ± 0.025 | 4.812 ± 0.039 |
4.607 ± 0.039 | 6.921 ± 0.035 |
4.888 ± 0.032 | 5.78 ± 0.041 |
1.128 ± 0.015 | 3.358 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
