
Simian foamy virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Ortervirales; Retroviridae; Spumaretrovirinae; Spumavirus
Average proteome isoelectric point is 7.08
Get precalculated fractions of proteins
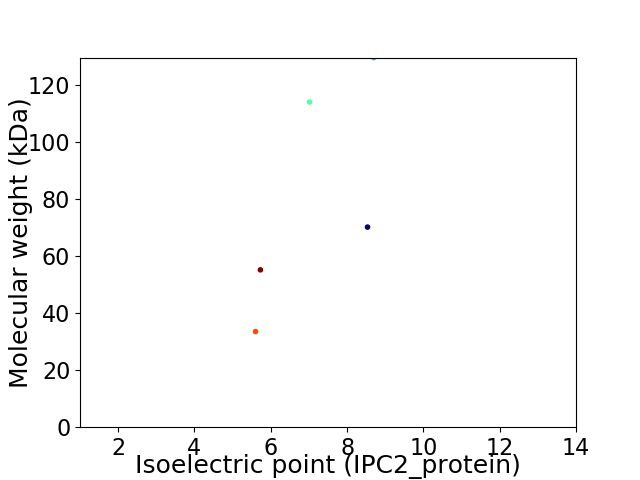
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|K7YNA6|K7YNA6_9RETR Pr125Pol OS=Simian foamy virus OX=11642 GN=pol PE=4 SV=1
MM1 pKa = 7.74DD2 pKa = 4.5TYY4 pKa = 11.16QEE6 pKa = 4.26EE7 pKa = 4.39EE8 pKa = 4.68SVASTSGVQDD18 pKa = 3.74LQTLSEE24 pKa = 4.24LVGPEE29 pKa = 3.58AAGEE33 pKa = 4.45GEE35 pKa = 4.49DD36 pKa = 3.77TLSDD40 pKa = 3.58TEE42 pKa = 4.35EE43 pKa = 3.84VSRR46 pKa = 11.84RR47 pKa = 11.84SRR49 pKa = 11.84KK50 pKa = 6.03NTKK53 pKa = 9.9RR54 pKa = 11.84GAKK57 pKa = 9.22MITFHH62 pKa = 6.89AYY64 pKa = 9.67KK65 pKa = 10.14EE66 pKa = 4.19IEE68 pKa = 4.38EE69 pKa = 4.67KK70 pKa = 11.0NPQNLKK76 pKa = 8.67LHH78 pKa = 5.78NWIPTPEE85 pKa = 4.05EE86 pKa = 3.8MSKK89 pKa = 10.65SICSKK94 pKa = 10.71FILHH98 pKa = 6.8GLYY101 pKa = 10.31SAEE104 pKa = 3.99KK105 pKa = 10.17VIEE108 pKa = 4.01ILNMPFEE115 pKa = 4.58ISWDD119 pKa = 3.6QSEE122 pKa = 4.6TNPKK126 pKa = 9.93CFIVSYY132 pKa = 9.15TCIYY136 pKa = 9.82CSAIIHH142 pKa = 6.24EE143 pKa = 4.74PMPVTYY149 pKa = 10.5NADD152 pKa = 3.17VGVWVKK158 pKa = 10.45LNPLRR163 pKa = 11.84GVVGSAVFMMKK174 pKa = 9.75KK175 pKa = 9.71HH176 pKa = 6.05EE177 pKa = 4.55KK178 pKa = 8.84NCCFIKK184 pKa = 10.42PSPSKK189 pKa = 10.88QEE191 pKa = 3.98GPKK194 pKa = 9.77PRR196 pKa = 11.84PRR198 pKa = 11.84HH199 pKa = 6.0DD200 pKa = 3.14PVLRR204 pKa = 11.84CKK206 pKa = 10.66AFEE209 pKa = 4.19RR210 pKa = 11.84NYY212 pKa = 10.47KK213 pKa = 9.62PRR215 pKa = 11.84PKK217 pKa = 10.02RR218 pKa = 11.84PRR220 pKa = 11.84PGPINNEE227 pKa = 3.9PCASSSDD234 pKa = 3.83PLAVQPGSLCTDD246 pKa = 4.16PVWDD250 pKa = 4.35PAAILQVLSEE260 pKa = 4.09QRR262 pKa = 11.84EE263 pKa = 4.22DD264 pKa = 3.69SQSLAIHH271 pKa = 6.11MSGGPFWEE279 pKa = 4.21EE280 pKa = 4.4VYY282 pKa = 10.84GDD284 pKa = 4.37SYY286 pKa = 10.61MGPPTRR292 pKa = 11.84PGDD295 pKa = 3.7HH296 pKa = 6.23TVLL299 pKa = 3.97
MM1 pKa = 7.74DD2 pKa = 4.5TYY4 pKa = 11.16QEE6 pKa = 4.26EE7 pKa = 4.39EE8 pKa = 4.68SVASTSGVQDD18 pKa = 3.74LQTLSEE24 pKa = 4.24LVGPEE29 pKa = 3.58AAGEE33 pKa = 4.45GEE35 pKa = 4.49DD36 pKa = 3.77TLSDD40 pKa = 3.58TEE42 pKa = 4.35EE43 pKa = 3.84VSRR46 pKa = 11.84RR47 pKa = 11.84SRR49 pKa = 11.84KK50 pKa = 6.03NTKK53 pKa = 9.9RR54 pKa = 11.84GAKK57 pKa = 9.22MITFHH62 pKa = 6.89AYY64 pKa = 9.67KK65 pKa = 10.14EE66 pKa = 4.19IEE68 pKa = 4.38EE69 pKa = 4.67KK70 pKa = 11.0NPQNLKK76 pKa = 8.67LHH78 pKa = 5.78NWIPTPEE85 pKa = 4.05EE86 pKa = 3.8MSKK89 pKa = 10.65SICSKK94 pKa = 10.71FILHH98 pKa = 6.8GLYY101 pKa = 10.31SAEE104 pKa = 3.99KK105 pKa = 10.17VIEE108 pKa = 4.01ILNMPFEE115 pKa = 4.58ISWDD119 pKa = 3.6QSEE122 pKa = 4.6TNPKK126 pKa = 9.93CFIVSYY132 pKa = 9.15TCIYY136 pKa = 9.82CSAIIHH142 pKa = 6.24EE143 pKa = 4.74PMPVTYY149 pKa = 10.5NADD152 pKa = 3.17VGVWVKK158 pKa = 10.45LNPLRR163 pKa = 11.84GVVGSAVFMMKK174 pKa = 9.75KK175 pKa = 9.71HH176 pKa = 6.05EE177 pKa = 4.55KK178 pKa = 8.84NCCFIKK184 pKa = 10.42PSPSKK189 pKa = 10.88QEE191 pKa = 3.98GPKK194 pKa = 9.77PRR196 pKa = 11.84PRR198 pKa = 11.84HH199 pKa = 6.0DD200 pKa = 3.14PVLRR204 pKa = 11.84CKK206 pKa = 10.66AFEE209 pKa = 4.19RR210 pKa = 11.84NYY212 pKa = 10.47KK213 pKa = 9.62PRR215 pKa = 11.84PKK217 pKa = 10.02RR218 pKa = 11.84PRR220 pKa = 11.84PGPINNEE227 pKa = 3.9PCASSSDD234 pKa = 3.83PLAVQPGSLCTDD246 pKa = 4.16PVWDD250 pKa = 4.35PAAILQVLSEE260 pKa = 4.09QRR262 pKa = 11.84EE263 pKa = 4.22DD264 pKa = 3.69SQSLAIHH271 pKa = 6.11MSGGPFWEE279 pKa = 4.21EE280 pKa = 4.4VYY282 pKa = 10.84GDD284 pKa = 4.37SYY286 pKa = 10.61MGPPTRR292 pKa = 11.84PGDD295 pKa = 3.7HH296 pKa = 6.23TVLL299 pKa = 3.97
Molecular weight: 33.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K7Z2A0|K7Z2A0_9RETR Gag polyprotein OS=Simian foamy virus OX=11642 GN=gag PE=4 SV=1
MM1 pKa = 7.58NPLQLLQPLQAEE13 pKa = 4.58VKK15 pKa = 9.03GNKK18 pKa = 9.55LIAHH22 pKa = 7.1WDD24 pKa = 3.32SGASITCIPEE34 pKa = 3.83SFLEE38 pKa = 4.23EE39 pKa = 3.66EE40 pKa = 4.48TPIKK44 pKa = 9.66KK45 pKa = 8.81TIIKK49 pKa = 8.36TIHH52 pKa = 5.28GQKK55 pKa = 8.18EE56 pKa = 3.93QKK58 pKa = 10.41VYY60 pKa = 11.34YY61 pKa = 9.47LTFKK65 pKa = 10.75VNGRR69 pKa = 11.84KK70 pKa = 9.15VEE72 pKa = 4.14AEE74 pKa = 4.44VIASPYY80 pKa = 10.8DD81 pKa = 3.83YY82 pKa = 10.89ILLSPTDD89 pKa = 4.01VPWLVQKK96 pKa = 10.19PLQLTILVPLQDD108 pKa = 3.74YY109 pKa = 9.76QSRR112 pKa = 11.84ILEE115 pKa = 4.13KK116 pKa = 9.68TALPEE121 pKa = 3.92EE122 pKa = 4.71FKK124 pKa = 11.13KK125 pKa = 10.53QLQTLFLKK133 pKa = 10.71YY134 pKa = 10.67DD135 pKa = 3.76NLWQHH140 pKa = 6.27WEE142 pKa = 3.98NQVGHH147 pKa = 6.44RR148 pKa = 11.84KK149 pKa = 8.88IRR151 pKa = 11.84PHH153 pKa = 6.76NIATGDD159 pKa = 3.71YY160 pKa = 9.61PPRR163 pKa = 11.84PQKK166 pKa = 10.35QYY168 pKa = 10.54PINPKK173 pKa = 9.76ARR175 pKa = 11.84SSIQVVIDD183 pKa = 4.03DD184 pKa = 4.51LLKK187 pKa = 10.94QGVLVQQNSTMNTPVYY203 pKa = 9.51PVPKK207 pKa = 9.66PDD209 pKa = 3.26GRR211 pKa = 11.84WRR213 pKa = 11.84MVLDD217 pKa = 3.37YY218 pKa = 11.29RR219 pKa = 11.84EE220 pKa = 4.13VNKK223 pKa = 9.58TIPLIAAQNQHH234 pKa = 5.64SAGILATIVRR244 pKa = 11.84KK245 pKa = 9.84KK246 pKa = 11.06YY247 pKa = 9.06KK248 pKa = 8.27TTLDD252 pKa = 3.57LANGFWAHH260 pKa = 7.43PITPEE265 pKa = 4.44SYY267 pKa = 9.5WLTAFTWQGKK277 pKa = 6.31QYY279 pKa = 10.3CWTRR283 pKa = 11.84LPQGFLNSPALFTADD298 pKa = 3.46VVDD301 pKa = 4.32LLKK304 pKa = 10.84EE305 pKa = 3.81ISNVQAYY312 pKa = 10.42VDD314 pKa = 4.91DD315 pKa = 5.2IYY317 pKa = 11.3LSHH320 pKa = 8.21DD321 pKa = 4.13DD322 pKa = 3.97PQEE325 pKa = 4.03HH326 pKa = 6.83LNQLEE331 pKa = 4.31KK332 pKa = 11.01VFQILLQAGYY342 pKa = 10.34VVSLKK347 pKa = 10.54KK348 pKa = 10.59SEE350 pKa = 4.12IAQKK354 pKa = 9.0TVEE357 pKa = 4.04FLGFNITKK365 pKa = 9.78EE366 pKa = 4.22GRR368 pKa = 11.84GLTEE372 pKa = 3.8AFKK375 pKa = 11.17AKK377 pKa = 10.37LLDD380 pKa = 3.27ITPPKK385 pKa = 10.15DD386 pKa = 3.48LKK388 pKa = 10.77QLQSILGLLNFARR401 pKa = 11.84NFILNFAEE409 pKa = 4.49LVKK412 pKa = 10.28PLYY415 pKa = 10.76SLISSAKK422 pKa = 8.61GKK424 pKa = 10.29YY425 pKa = 9.43IEE427 pKa = 4.27WSNEE431 pKa = 3.35NTVQLQTIIKK441 pKa = 9.62ALNDD445 pKa = 3.51ADD447 pKa = 3.95NLEE450 pKa = 3.86EE451 pKa = 4.93RR452 pKa = 11.84IPEE455 pKa = 3.62KK456 pKa = 10.77RR457 pKa = 11.84LIIKK461 pKa = 10.12VNTSPSAGYY470 pKa = 9.09VRR472 pKa = 11.84YY473 pKa = 10.45YY474 pKa = 11.15NEE476 pKa = 4.16TGKK479 pKa = 10.81KK480 pKa = 9.46PIMYY484 pKa = 9.87LNYY487 pKa = 9.51VFSKK491 pKa = 11.38AEE493 pKa = 3.87LKK495 pKa = 9.27FTLLEE500 pKa = 4.21KK501 pKa = 10.93LLTTMHH507 pKa = 6.98KK508 pKa = 10.82ALIKK512 pKa = 11.07AMDD515 pKa = 3.95LAMGQEE521 pKa = 3.94ILVYY525 pKa = 10.3SPVVSMTKK533 pKa = 9.45IQKK536 pKa = 8.43TPIPEE541 pKa = 4.03RR542 pKa = 11.84KK543 pKa = 9.23ALPIRR548 pKa = 11.84WITWMTYY555 pKa = 10.7LEE557 pKa = 4.43DD558 pKa = 3.7PRR560 pKa = 11.84IQFHH564 pKa = 5.99YY565 pKa = 10.92DD566 pKa = 2.7KK567 pKa = 10.67TLPEE571 pKa = 4.72LKK573 pKa = 10.38NIPDD577 pKa = 3.61VLTEE581 pKa = 3.97NSSKK585 pKa = 10.89IMIHH589 pKa = 6.1PSQYY593 pKa = 10.99NSVFYY598 pKa = 10.53TDD600 pKa = 3.69GSAIRR605 pKa = 11.84SPDD608 pKa = 3.09PTKK611 pKa = 10.91SHH613 pKa = 6.21NAGMGIVQAKK623 pKa = 8.7FSPEE627 pKa = 3.79LQVINQWSIPLGNHH641 pKa = 5.23TAQMAEE647 pKa = 3.6IAAVEE652 pKa = 4.32FACKK656 pKa = 9.73KK657 pKa = 9.7ALKK660 pKa = 8.67ITGPILIITDD670 pKa = 3.6SFYY673 pKa = 11.35VAEE676 pKa = 4.48SANKK680 pKa = 8.91EE681 pKa = 3.85LPYY684 pKa = 9.92WKK686 pKa = 10.68SNGFVNNKK694 pKa = 9.3KK695 pKa = 10.41KK696 pKa = 10.05PLKK699 pKa = 10.29HH700 pKa = 5.03VSKK703 pKa = 9.37WKK705 pKa = 10.68SIAEE709 pKa = 4.16CLSLKK714 pKa = 10.35PDD716 pKa = 2.92ITIQHH721 pKa = 5.9EE722 pKa = 4.27RR723 pKa = 11.84GHH725 pKa = 5.48QPIYY729 pKa = 10.56TSIHH733 pKa = 5.42TEE735 pKa = 4.02GNALADD741 pKa = 3.72KK742 pKa = 10.67LATQGSYY749 pKa = 9.19VVNNNDD755 pKa = 4.09KK756 pKa = 10.94KK757 pKa = 10.69PNLDD761 pKa = 3.68AEE763 pKa = 4.7LDD765 pKa = 3.58HH766 pKa = 7.08LIQGKK771 pKa = 8.1YY772 pKa = 8.64PKK774 pKa = 9.73GYY776 pKa = 8.36PKK778 pKa = 10.2QYY780 pKa = 9.44PYY782 pKa = 11.62YY783 pKa = 9.86MEE785 pKa = 5.19DD786 pKa = 2.9GKK788 pKa = 11.64VKK790 pKa = 10.32VNRR793 pKa = 11.84PEE795 pKa = 3.86GTKK798 pKa = 9.91IIPPSLEE805 pKa = 3.54RR806 pKa = 11.84AGIVQKK812 pKa = 10.91AHH814 pKa = 6.18NLAHH818 pKa = 6.88TGRR821 pKa = 11.84EE822 pKa = 3.94ATLLKK827 pKa = 10.2IANLYY832 pKa = 6.99WWPNMRR838 pKa = 11.84KK839 pKa = 9.79DD840 pKa = 3.63VVKK843 pKa = 10.73QLGRR847 pKa = 11.84CQQCLVTNAFNQTSGPILRR866 pKa = 11.84PTRR869 pKa = 11.84PLKK872 pKa = 10.76PFDD875 pKa = 4.3KK876 pKa = 10.94FFIDD880 pKa = 4.29YY881 pKa = 10.24IGPLPPSNGYY891 pKa = 8.92LHH893 pKa = 6.42VLVVVDD899 pKa = 4.43SMTGFTWLYY908 pKa = 5.79PTKK911 pKa = 10.77APTTNATVKK920 pKa = 10.39ALNVLTSIAVPKK932 pKa = 10.18VIHH935 pKa = 6.68SDD937 pKa = 2.8QGAAFTSSTFADD949 pKa = 3.46WAKK952 pKa = 10.69EE953 pKa = 3.63RR954 pKa = 11.84GIQLEE959 pKa = 4.34FSTPYY964 pKa = 10.25HH965 pKa = 5.98PQSSGKK971 pKa = 8.3VEE973 pKa = 4.3RR974 pKa = 11.84KK975 pKa = 10.0NSDD978 pKa = 2.61IKK980 pKa = 11.08RR981 pKa = 11.84LLTKK985 pKa = 10.58LLVGRR990 pKa = 11.84PTKK993 pKa = 10.03WYY995 pKa = 9.91DD996 pKa = 3.56LLPVVQLALNNSYY1009 pKa = 10.92SPSLKK1014 pKa = 9.01HH1015 pKa = 6.12TPHH1018 pKa = 6.28QLLFGIDD1025 pKa = 3.66SNTPFANQDD1034 pKa = 3.49TLDD1037 pKa = 3.66LTRR1040 pKa = 11.84EE1041 pKa = 4.18EE1042 pKa = 4.48EE1043 pKa = 4.13LSLLQEE1049 pKa = 4.45IRR1051 pKa = 11.84SSLHH1055 pKa = 5.69QPSTPPASTRR1065 pKa = 11.84SWSPIVGQIVQEE1077 pKa = 4.17RR1078 pKa = 11.84VPRR1081 pKa = 11.84PASLRR1086 pKa = 11.84PRR1088 pKa = 11.84WHH1090 pKa = 6.07KK1091 pKa = 9.93PSRR1094 pKa = 11.84IVDD1097 pKa = 3.34ILNEE1101 pKa = 4.07RR1102 pKa = 11.84TVVIVDD1108 pKa = 3.11HH1109 pKa = 6.75LGNNRR1114 pKa = 11.84TVSIDD1119 pKa = 3.5NLKK1122 pKa = 8.92LTPHH1126 pKa = 6.48QDD1128 pKa = 2.79GTSNVSSTMDD1138 pKa = 3.22HH1139 pKa = 7.04LEE1141 pKa = 3.79
MM1 pKa = 7.58NPLQLLQPLQAEE13 pKa = 4.58VKK15 pKa = 9.03GNKK18 pKa = 9.55LIAHH22 pKa = 7.1WDD24 pKa = 3.32SGASITCIPEE34 pKa = 3.83SFLEE38 pKa = 4.23EE39 pKa = 3.66EE40 pKa = 4.48TPIKK44 pKa = 9.66KK45 pKa = 8.81TIIKK49 pKa = 8.36TIHH52 pKa = 5.28GQKK55 pKa = 8.18EE56 pKa = 3.93QKK58 pKa = 10.41VYY60 pKa = 11.34YY61 pKa = 9.47LTFKK65 pKa = 10.75VNGRR69 pKa = 11.84KK70 pKa = 9.15VEE72 pKa = 4.14AEE74 pKa = 4.44VIASPYY80 pKa = 10.8DD81 pKa = 3.83YY82 pKa = 10.89ILLSPTDD89 pKa = 4.01VPWLVQKK96 pKa = 10.19PLQLTILVPLQDD108 pKa = 3.74YY109 pKa = 9.76QSRR112 pKa = 11.84ILEE115 pKa = 4.13KK116 pKa = 9.68TALPEE121 pKa = 3.92EE122 pKa = 4.71FKK124 pKa = 11.13KK125 pKa = 10.53QLQTLFLKK133 pKa = 10.71YY134 pKa = 10.67DD135 pKa = 3.76NLWQHH140 pKa = 6.27WEE142 pKa = 3.98NQVGHH147 pKa = 6.44RR148 pKa = 11.84KK149 pKa = 8.88IRR151 pKa = 11.84PHH153 pKa = 6.76NIATGDD159 pKa = 3.71YY160 pKa = 9.61PPRR163 pKa = 11.84PQKK166 pKa = 10.35QYY168 pKa = 10.54PINPKK173 pKa = 9.76ARR175 pKa = 11.84SSIQVVIDD183 pKa = 4.03DD184 pKa = 4.51LLKK187 pKa = 10.94QGVLVQQNSTMNTPVYY203 pKa = 9.51PVPKK207 pKa = 9.66PDD209 pKa = 3.26GRR211 pKa = 11.84WRR213 pKa = 11.84MVLDD217 pKa = 3.37YY218 pKa = 11.29RR219 pKa = 11.84EE220 pKa = 4.13VNKK223 pKa = 9.58TIPLIAAQNQHH234 pKa = 5.64SAGILATIVRR244 pKa = 11.84KK245 pKa = 9.84KK246 pKa = 11.06YY247 pKa = 9.06KK248 pKa = 8.27TTLDD252 pKa = 3.57LANGFWAHH260 pKa = 7.43PITPEE265 pKa = 4.44SYY267 pKa = 9.5WLTAFTWQGKK277 pKa = 6.31QYY279 pKa = 10.3CWTRR283 pKa = 11.84LPQGFLNSPALFTADD298 pKa = 3.46VVDD301 pKa = 4.32LLKK304 pKa = 10.84EE305 pKa = 3.81ISNVQAYY312 pKa = 10.42VDD314 pKa = 4.91DD315 pKa = 5.2IYY317 pKa = 11.3LSHH320 pKa = 8.21DD321 pKa = 4.13DD322 pKa = 3.97PQEE325 pKa = 4.03HH326 pKa = 6.83LNQLEE331 pKa = 4.31KK332 pKa = 11.01VFQILLQAGYY342 pKa = 10.34VVSLKK347 pKa = 10.54KK348 pKa = 10.59SEE350 pKa = 4.12IAQKK354 pKa = 9.0TVEE357 pKa = 4.04FLGFNITKK365 pKa = 9.78EE366 pKa = 4.22GRR368 pKa = 11.84GLTEE372 pKa = 3.8AFKK375 pKa = 11.17AKK377 pKa = 10.37LLDD380 pKa = 3.27ITPPKK385 pKa = 10.15DD386 pKa = 3.48LKK388 pKa = 10.77QLQSILGLLNFARR401 pKa = 11.84NFILNFAEE409 pKa = 4.49LVKK412 pKa = 10.28PLYY415 pKa = 10.76SLISSAKK422 pKa = 8.61GKK424 pKa = 10.29YY425 pKa = 9.43IEE427 pKa = 4.27WSNEE431 pKa = 3.35NTVQLQTIIKK441 pKa = 9.62ALNDD445 pKa = 3.51ADD447 pKa = 3.95NLEE450 pKa = 3.86EE451 pKa = 4.93RR452 pKa = 11.84IPEE455 pKa = 3.62KK456 pKa = 10.77RR457 pKa = 11.84LIIKK461 pKa = 10.12VNTSPSAGYY470 pKa = 9.09VRR472 pKa = 11.84YY473 pKa = 10.45YY474 pKa = 11.15NEE476 pKa = 4.16TGKK479 pKa = 10.81KK480 pKa = 9.46PIMYY484 pKa = 9.87LNYY487 pKa = 9.51VFSKK491 pKa = 11.38AEE493 pKa = 3.87LKK495 pKa = 9.27FTLLEE500 pKa = 4.21KK501 pKa = 10.93LLTTMHH507 pKa = 6.98KK508 pKa = 10.82ALIKK512 pKa = 11.07AMDD515 pKa = 3.95LAMGQEE521 pKa = 3.94ILVYY525 pKa = 10.3SPVVSMTKK533 pKa = 9.45IQKK536 pKa = 8.43TPIPEE541 pKa = 4.03RR542 pKa = 11.84KK543 pKa = 9.23ALPIRR548 pKa = 11.84WITWMTYY555 pKa = 10.7LEE557 pKa = 4.43DD558 pKa = 3.7PRR560 pKa = 11.84IQFHH564 pKa = 5.99YY565 pKa = 10.92DD566 pKa = 2.7KK567 pKa = 10.67TLPEE571 pKa = 4.72LKK573 pKa = 10.38NIPDD577 pKa = 3.61VLTEE581 pKa = 3.97NSSKK585 pKa = 10.89IMIHH589 pKa = 6.1PSQYY593 pKa = 10.99NSVFYY598 pKa = 10.53TDD600 pKa = 3.69GSAIRR605 pKa = 11.84SPDD608 pKa = 3.09PTKK611 pKa = 10.91SHH613 pKa = 6.21NAGMGIVQAKK623 pKa = 8.7FSPEE627 pKa = 3.79LQVINQWSIPLGNHH641 pKa = 5.23TAQMAEE647 pKa = 3.6IAAVEE652 pKa = 4.32FACKK656 pKa = 9.73KK657 pKa = 9.7ALKK660 pKa = 8.67ITGPILIITDD670 pKa = 3.6SFYY673 pKa = 11.35VAEE676 pKa = 4.48SANKK680 pKa = 8.91EE681 pKa = 3.85LPYY684 pKa = 9.92WKK686 pKa = 10.68SNGFVNNKK694 pKa = 9.3KK695 pKa = 10.41KK696 pKa = 10.05PLKK699 pKa = 10.29HH700 pKa = 5.03VSKK703 pKa = 9.37WKK705 pKa = 10.68SIAEE709 pKa = 4.16CLSLKK714 pKa = 10.35PDD716 pKa = 2.92ITIQHH721 pKa = 5.9EE722 pKa = 4.27RR723 pKa = 11.84GHH725 pKa = 5.48QPIYY729 pKa = 10.56TSIHH733 pKa = 5.42TEE735 pKa = 4.02GNALADD741 pKa = 3.72KK742 pKa = 10.67LATQGSYY749 pKa = 9.19VVNNNDD755 pKa = 4.09KK756 pKa = 10.94KK757 pKa = 10.69PNLDD761 pKa = 3.68AEE763 pKa = 4.7LDD765 pKa = 3.58HH766 pKa = 7.08LIQGKK771 pKa = 8.1YY772 pKa = 8.64PKK774 pKa = 9.73GYY776 pKa = 8.36PKK778 pKa = 10.2QYY780 pKa = 9.44PYY782 pKa = 11.62YY783 pKa = 9.86MEE785 pKa = 5.19DD786 pKa = 2.9GKK788 pKa = 11.64VKK790 pKa = 10.32VNRR793 pKa = 11.84PEE795 pKa = 3.86GTKK798 pKa = 9.91IIPPSLEE805 pKa = 3.54RR806 pKa = 11.84AGIVQKK812 pKa = 10.91AHH814 pKa = 6.18NLAHH818 pKa = 6.88TGRR821 pKa = 11.84EE822 pKa = 3.94ATLLKK827 pKa = 10.2IANLYY832 pKa = 6.99WWPNMRR838 pKa = 11.84KK839 pKa = 9.79DD840 pKa = 3.63VVKK843 pKa = 10.73QLGRR847 pKa = 11.84CQQCLVTNAFNQTSGPILRR866 pKa = 11.84PTRR869 pKa = 11.84PLKK872 pKa = 10.76PFDD875 pKa = 4.3KK876 pKa = 10.94FFIDD880 pKa = 4.29YY881 pKa = 10.24IGPLPPSNGYY891 pKa = 8.92LHH893 pKa = 6.42VLVVVDD899 pKa = 4.43SMTGFTWLYY908 pKa = 5.79PTKK911 pKa = 10.77APTTNATVKK920 pKa = 10.39ALNVLTSIAVPKK932 pKa = 10.18VIHH935 pKa = 6.68SDD937 pKa = 2.8QGAAFTSSTFADD949 pKa = 3.46WAKK952 pKa = 10.69EE953 pKa = 3.63RR954 pKa = 11.84GIQLEE959 pKa = 4.34FSTPYY964 pKa = 10.25HH965 pKa = 5.98PQSSGKK971 pKa = 8.3VEE973 pKa = 4.3RR974 pKa = 11.84KK975 pKa = 10.0NSDD978 pKa = 2.61IKK980 pKa = 11.08RR981 pKa = 11.84LLTKK985 pKa = 10.58LLVGRR990 pKa = 11.84PTKK993 pKa = 10.03WYY995 pKa = 9.91DD996 pKa = 3.56LLPVVQLALNNSYY1009 pKa = 10.92SPSLKK1014 pKa = 9.01HH1015 pKa = 6.12TPHH1018 pKa = 6.28QLLFGIDD1025 pKa = 3.66SNTPFANQDD1034 pKa = 3.49TLDD1037 pKa = 3.66LTRR1040 pKa = 11.84EE1041 pKa = 4.18EE1042 pKa = 4.48EE1043 pKa = 4.13LSLLQEE1049 pKa = 4.45IRR1051 pKa = 11.84SSLHH1055 pKa = 5.69QPSTPPASTRR1065 pKa = 11.84SWSPIVGQIVQEE1077 pKa = 4.17RR1078 pKa = 11.84VPRR1081 pKa = 11.84PASLRR1086 pKa = 11.84PRR1088 pKa = 11.84WHH1090 pKa = 6.07KK1091 pKa = 9.93PSRR1094 pKa = 11.84IVDD1097 pKa = 3.34ILNEE1101 pKa = 4.07RR1102 pKa = 11.84TVVIVDD1108 pKa = 3.11HH1109 pKa = 6.75LGNNRR1114 pKa = 11.84TVSIDD1119 pKa = 3.5NLKK1122 pKa = 8.92LTPHH1126 pKa = 6.48QDD1128 pKa = 2.79GTSNVSSTMDD1138 pKa = 3.22HH1139 pKa = 7.04LEE1141 pKa = 3.79
Molecular weight: 129.59 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3553 |
299 |
1141 |
710.6 |
80.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.685 ± 0.185 | 1.379 ± 0.483 |
4.391 ± 0.21 | 6.023 ± 0.577 |
2.702 ± 0.176 | 5.573 ± 0.951 |
2.477 ± 0.236 | 6.642 ± 0.632 |
5.826 ± 1.276 | 9.063 ± 0.879 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.97 ± 0.234 | 5.094 ± 0.215 |
7.233 ± 0.839 | 5.77 ± 0.52 |
5.207 ± 0.939 | 6.98 ± 0.605 |
5.995 ± 0.28 | 6.164 ± 0.14 |
1.998 ± 0.272 | 3.828 ± 0.41 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
