
Yerba mate chlorosis-associated virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Betarhabdovirinae; Cytorhabdovirus; Yerba mate chlorosis-associated cytorhabdovirus
Average proteome isoelectric point is 7.22
Get precalculated fractions of proteins
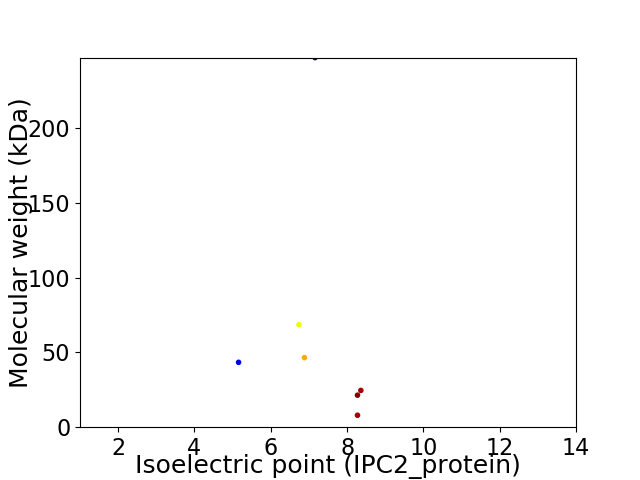
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W5S0M4|A0A1W5S0M4_9RHAB Large structural protein OS=Yerba mate chlorosis-associated virus OX=2487100 PE=4 SV=1
MM1 pKa = 7.67SNGFEE6 pKa = 4.14FKK8 pKa = 10.61KK9 pKa = 10.54IEE11 pKa = 4.17GKK13 pKa = 10.36LDD15 pKa = 3.52YY16 pKa = 11.19SGVQHH21 pKa = 6.39MSSALDD27 pKa = 4.06DD28 pKa = 5.12DD29 pKa = 5.74NMFADD34 pKa = 4.35KK35 pKa = 10.85NVSLEE40 pKa = 4.07NKK42 pKa = 10.29EE43 pKa = 4.42EE44 pKa = 4.21GTKK47 pKa = 8.94TAEE50 pKa = 3.82IVVTQNIPAASKK62 pKa = 10.26EE63 pKa = 4.26KK64 pKa = 10.73NSPFLKK70 pKa = 10.51ALSKK74 pKa = 9.97KK75 pKa = 9.17TEE77 pKa = 4.47DD78 pKa = 3.42NTGKK82 pKa = 10.28SGEE85 pKa = 4.21VEE87 pKa = 3.94EE88 pKa = 5.1SKK90 pKa = 9.87TPKK93 pKa = 9.89DD94 pKa = 3.01TDD96 pKa = 3.4KK97 pKa = 11.61AKK99 pKa = 8.69EE100 pKa = 4.25TKK102 pKa = 9.81SVKK105 pKa = 9.95SLKK108 pKa = 10.4EE109 pKa = 3.75NLKK112 pKa = 10.73SRR114 pKa = 11.84LEE116 pKa = 4.0DD117 pKa = 3.23HH118 pKa = 7.35LGRR121 pKa = 11.84SSNNSKK127 pKa = 10.9EE128 pKa = 4.04EE129 pKa = 4.02NKK131 pKa = 10.42DD132 pKa = 3.26SKK134 pKa = 11.32AVEE137 pKa = 4.08KK138 pKa = 10.12TKK140 pKa = 11.19KK141 pKa = 10.29NLSKK145 pKa = 10.88DD146 pKa = 3.43LNKK149 pKa = 9.96VLEE152 pKa = 4.35EE153 pKa = 4.29DD154 pKa = 4.2EE155 pKa = 4.87EE156 pKa = 5.36IEE158 pKa = 5.03DD159 pKa = 3.67EE160 pKa = 4.46SEE162 pKa = 4.0EE163 pKa = 4.1LFSPEE168 pKa = 4.77EE169 pKa = 3.81IRR171 pKa = 11.84DD172 pKa = 4.06CLDD175 pKa = 3.51LFCNKK180 pKa = 10.11LGVDD184 pKa = 5.23GEE186 pKa = 4.82SAWYY190 pKa = 9.02NHH192 pKa = 5.99FVASLNINGAVFSSEE207 pKa = 3.66IEE209 pKa = 4.15SWVSAIKK216 pKa = 10.42HH217 pKa = 4.7EE218 pKa = 4.37RR219 pKa = 11.84ATASHH224 pKa = 6.63RR225 pKa = 11.84AIKK228 pKa = 10.3EE229 pKa = 3.42ILTNLNTQVVRR240 pKa = 11.84LTAQVKK246 pKa = 10.11ALSDD250 pKa = 3.93TNGSLAKK257 pKa = 10.44QNEE260 pKa = 4.55SLLNMVDD267 pKa = 3.84SMRR270 pKa = 11.84KK271 pKa = 8.04NQEE274 pKa = 3.13EE275 pKa = 4.29WLNTQLEE282 pKa = 4.39LRR284 pKa = 11.84QGAIRR289 pKa = 11.84GGYY292 pKa = 8.3QKK294 pKa = 10.93EE295 pKa = 4.14KK296 pKa = 10.99GKK298 pKa = 8.89LTEE301 pKa = 4.17EE302 pKa = 4.24MSDD305 pKa = 3.56EE306 pKa = 4.23NASSGSYY313 pKa = 10.72YY314 pKa = 10.62LDD316 pKa = 3.4EE317 pKa = 4.99TDD319 pKa = 5.95DD320 pKa = 3.82EE321 pKa = 4.61TSEE324 pKa = 4.51LLVSHH329 pKa = 7.39FLTLLKK335 pKa = 10.47INSTQIQDD343 pKa = 3.3PRR345 pKa = 11.84VRR347 pKa = 11.84MAVRR351 pKa = 11.84SLFPMKK357 pKa = 10.19ILVAACEE364 pKa = 4.0HH365 pKa = 7.29GIRR368 pKa = 11.84EE369 pKa = 3.98ADD371 pKa = 3.41KK372 pKa = 10.94PKK374 pKa = 10.73ALEE377 pKa = 4.32KK378 pKa = 10.49LKK380 pKa = 10.81EE381 pKa = 4.14WILNNN386 pKa = 3.72
MM1 pKa = 7.67SNGFEE6 pKa = 4.14FKK8 pKa = 10.61KK9 pKa = 10.54IEE11 pKa = 4.17GKK13 pKa = 10.36LDD15 pKa = 3.52YY16 pKa = 11.19SGVQHH21 pKa = 6.39MSSALDD27 pKa = 4.06DD28 pKa = 5.12DD29 pKa = 5.74NMFADD34 pKa = 4.35KK35 pKa = 10.85NVSLEE40 pKa = 4.07NKK42 pKa = 10.29EE43 pKa = 4.42EE44 pKa = 4.21GTKK47 pKa = 8.94TAEE50 pKa = 3.82IVVTQNIPAASKK62 pKa = 10.26EE63 pKa = 4.26KK64 pKa = 10.73NSPFLKK70 pKa = 10.51ALSKK74 pKa = 9.97KK75 pKa = 9.17TEE77 pKa = 4.47DD78 pKa = 3.42NTGKK82 pKa = 10.28SGEE85 pKa = 4.21VEE87 pKa = 3.94EE88 pKa = 5.1SKK90 pKa = 9.87TPKK93 pKa = 9.89DD94 pKa = 3.01TDD96 pKa = 3.4KK97 pKa = 11.61AKK99 pKa = 8.69EE100 pKa = 4.25TKK102 pKa = 9.81SVKK105 pKa = 9.95SLKK108 pKa = 10.4EE109 pKa = 3.75NLKK112 pKa = 10.73SRR114 pKa = 11.84LEE116 pKa = 4.0DD117 pKa = 3.23HH118 pKa = 7.35LGRR121 pKa = 11.84SSNNSKK127 pKa = 10.9EE128 pKa = 4.04EE129 pKa = 4.02NKK131 pKa = 10.42DD132 pKa = 3.26SKK134 pKa = 11.32AVEE137 pKa = 4.08KK138 pKa = 10.12TKK140 pKa = 11.19KK141 pKa = 10.29NLSKK145 pKa = 10.88DD146 pKa = 3.43LNKK149 pKa = 9.96VLEE152 pKa = 4.35EE153 pKa = 4.29DD154 pKa = 4.2EE155 pKa = 4.87EE156 pKa = 5.36IEE158 pKa = 5.03DD159 pKa = 3.67EE160 pKa = 4.46SEE162 pKa = 4.0EE163 pKa = 4.1LFSPEE168 pKa = 4.77EE169 pKa = 3.81IRR171 pKa = 11.84DD172 pKa = 4.06CLDD175 pKa = 3.51LFCNKK180 pKa = 10.11LGVDD184 pKa = 5.23GEE186 pKa = 4.82SAWYY190 pKa = 9.02NHH192 pKa = 5.99FVASLNINGAVFSSEE207 pKa = 3.66IEE209 pKa = 4.15SWVSAIKK216 pKa = 10.42HH217 pKa = 4.7EE218 pKa = 4.37RR219 pKa = 11.84ATASHH224 pKa = 6.63RR225 pKa = 11.84AIKK228 pKa = 10.3EE229 pKa = 3.42ILTNLNTQVVRR240 pKa = 11.84LTAQVKK246 pKa = 10.11ALSDD250 pKa = 3.93TNGSLAKK257 pKa = 10.44QNEE260 pKa = 4.55SLLNMVDD267 pKa = 3.84SMRR270 pKa = 11.84KK271 pKa = 8.04NQEE274 pKa = 3.13EE275 pKa = 4.29WLNTQLEE282 pKa = 4.39LRR284 pKa = 11.84QGAIRR289 pKa = 11.84GGYY292 pKa = 8.3QKK294 pKa = 10.93EE295 pKa = 4.14KK296 pKa = 10.99GKK298 pKa = 8.89LTEE301 pKa = 4.17EE302 pKa = 4.24MSDD305 pKa = 3.56EE306 pKa = 4.23NASSGSYY313 pKa = 10.72YY314 pKa = 10.62LDD316 pKa = 3.4EE317 pKa = 4.99TDD319 pKa = 5.95DD320 pKa = 3.82EE321 pKa = 4.61TSEE324 pKa = 4.51LLVSHH329 pKa = 7.39FLTLLKK335 pKa = 10.47INSTQIQDD343 pKa = 3.3PRR345 pKa = 11.84VRR347 pKa = 11.84MAVRR351 pKa = 11.84SLFPMKK357 pKa = 10.19ILVAACEE364 pKa = 4.0HH365 pKa = 7.29GIRR368 pKa = 11.84EE369 pKa = 3.98ADD371 pKa = 3.41KK372 pKa = 10.94PKK374 pKa = 10.73ALEE377 pKa = 4.32KK378 pKa = 10.49LKK380 pKa = 10.81EE381 pKa = 4.14WILNNN386 pKa = 3.72
Molecular weight: 43.4 kDa
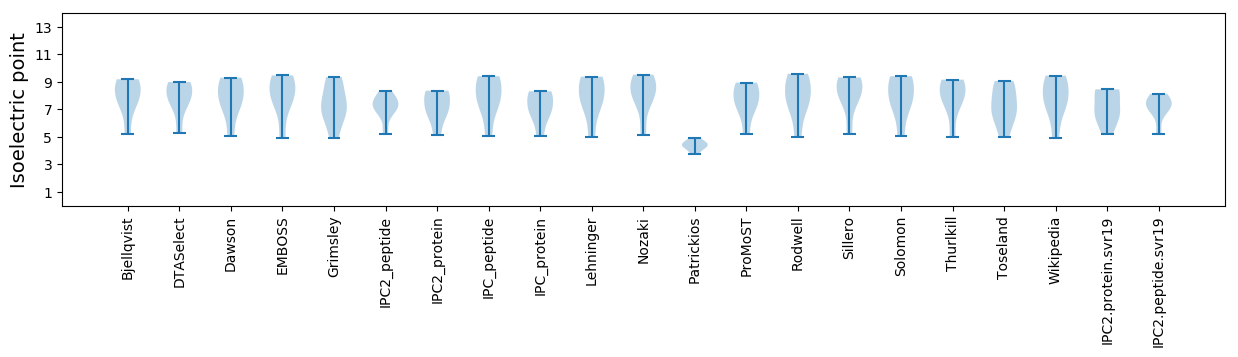
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W5RYH2|A0A1W5RYH2_9RHAB Phosphoprotein OS=Yerba mate chlorosis-associated virus OX=2487100 PE=4 SV=1
MM1 pKa = 7.24FFIIKK6 pKa = 9.58PLIKK10 pKa = 10.52LLVYY14 pKa = 8.9LLCLFFLLSVLVWGCIMMCDD34 pKa = 5.12LYY36 pKa = 11.18EE37 pKa = 4.06CHH39 pKa = 7.0NNLKK43 pKa = 10.84LNIISMKK50 pKa = 10.6KK51 pKa = 10.57FIDD54 pKa = 3.73CVKK57 pKa = 10.45KK58 pKa = 10.32KK59 pKa = 10.07RR60 pKa = 11.84QNPFCPLL67 pKa = 3.2
MM1 pKa = 7.24FFIIKK6 pKa = 9.58PLIKK10 pKa = 10.52LLVYY14 pKa = 8.9LLCLFFLLSVLVWGCIMMCDD34 pKa = 5.12LYY36 pKa = 11.18EE37 pKa = 4.06CHH39 pKa = 7.0NNLKK43 pKa = 10.84LNIISMKK50 pKa = 10.6KK51 pKa = 10.57FIDD54 pKa = 3.73CVKK57 pKa = 10.45KK58 pKa = 10.32KK59 pKa = 10.07RR60 pKa = 11.84QNPFCPLL67 pKa = 3.2
Molecular weight: 8.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4015 |
67 |
2134 |
573.6 |
65.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.359 ± 0.752 | 1.893 ± 0.451 |
5.355 ± 0.496 | 6.426 ± 0.972 |
4.633 ± 0.452 | 5.056 ± 0.256 |
2.391 ± 0.297 | 7.547 ± 0.72 |
8.02 ± 0.818 | 10.66 ± 0.439 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.64 ± 0.291 | 5.305 ± 0.448 |
4.209 ± 0.472 | 3.362 ± 0.426 |
3.985 ± 0.596 | 8.344 ± 0.741 |
5.28 ± 0.766 | 5.006 ± 0.234 |
1.694 ± 0.211 | 3.836 ± 0.569 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
