
Faecalibacterium phage FP_Taranis
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Taranisvirus; Faecalibacterium virus Taranis
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
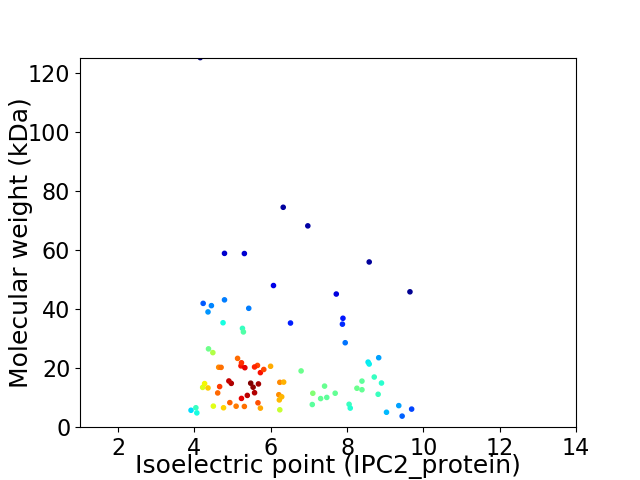
Virtual 2D-PAGE plot for 85 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
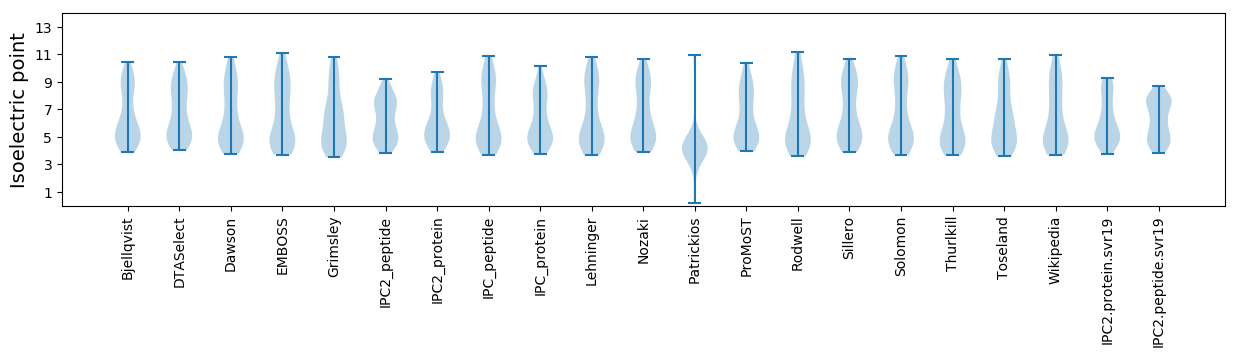
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2K9V452|A0A2K9V452_9CAUD Portal protein OS=Faecalibacterium phage FP_Taranis OX=2070186 PE=4 SV=1
MM1 pKa = 6.8NTVTVYY7 pKa = 10.81ACPSVPMDD15 pKa = 4.76SIEE18 pKa = 4.16CTIEE22 pKa = 3.51YY23 pKa = 10.64DD24 pKa = 3.74PAVVEE29 pKa = 5.05AFLHH33 pKa = 6.12PPNSGQEE40 pKa = 3.99RR41 pKa = 11.84ADD43 pKa = 3.94DD44 pKa = 4.2GSFGDD49 pKa = 4.12EE50 pKa = 3.96KK51 pKa = 11.53VLL53 pKa = 4.34
MM1 pKa = 6.8NTVTVYY7 pKa = 10.81ACPSVPMDD15 pKa = 4.76SIEE18 pKa = 4.16CTIEE22 pKa = 3.51YY23 pKa = 10.64DD24 pKa = 3.74PAVVEE29 pKa = 5.05AFLHH33 pKa = 6.12PPNSGQEE40 pKa = 3.99RR41 pKa = 11.84ADD43 pKa = 3.94DD44 pKa = 4.2GSFGDD49 pKa = 4.12EE50 pKa = 3.96KK51 pKa = 11.53VLL53 pKa = 4.34
Molecular weight: 5.75 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2K9V475|A0A2K9V475_9CAUD Uncharacterized protein OS=Faecalibacterium phage FP_Taranis OX=2070186 PE=4 SV=1
MM1 pKa = 7.4TSQEE5 pKa = 3.83RR6 pKa = 11.84HH7 pKa = 4.26EE8 pKa = 4.47ARR10 pKa = 11.84YY11 pKa = 8.6QRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84AARR18 pKa = 11.84RR19 pKa = 11.84ARR21 pKa = 11.84QEE23 pKa = 3.75ARR25 pKa = 11.84CAALGSLGEE34 pKa = 4.26VFSYY38 pKa = 8.7HH39 pKa = 6.0TMFKK43 pKa = 10.6YY44 pKa = 10.39GRR46 pKa = 11.84KK47 pKa = 8.33CCNGIRR53 pKa = 11.84WKK55 pKa = 10.81QSTQNFEE62 pKa = 3.85RR63 pKa = 11.84HH64 pKa = 5.69LFSHH68 pKa = 6.08TAKK71 pKa = 9.83QRR73 pKa = 11.84RR74 pKa = 11.84LILAKK79 pKa = 9.61RR80 pKa = 11.84WRR82 pKa = 11.84PKK84 pKa = 10.35KK85 pKa = 9.19YY86 pKa = 9.97VHH88 pKa = 5.94FTVCEE93 pKa = 3.79RR94 pKa = 11.84GKK96 pKa = 10.3IRR98 pKa = 11.84GIDD101 pKa = 3.48APHH104 pKa = 5.75ITDD107 pKa = 3.64RR108 pKa = 11.84QIHH111 pKa = 5.31KK112 pKa = 10.37VISKK116 pKa = 10.21EE117 pKa = 4.0VLEE120 pKa = 4.27PLYY123 pKa = 10.31DD124 pKa = 3.52PSMIYY129 pKa = 10.77DD130 pKa = 3.68NGASRR135 pKa = 11.84IGKK138 pKa = 8.72GLHH141 pKa = 4.59WQIKK145 pKa = 9.66RR146 pKa = 11.84IKK148 pKa = 8.76QQLARR153 pKa = 11.84HH154 pKa = 4.47YY155 pKa = 9.83RR156 pKa = 11.84KK157 pKa = 9.73YY158 pKa = 10.87GRR160 pKa = 11.84AGGVLLLDD168 pKa = 4.04LKK170 pKa = 11.15KK171 pKa = 10.3FFPYY175 pKa = 10.15APHH178 pKa = 6.43SIIYY182 pKa = 9.29QRR184 pKa = 11.84HH185 pKa = 3.56QRR187 pKa = 11.84YY188 pKa = 8.88ILNPDD193 pKa = 3.54FRR195 pKa = 11.84RR196 pKa = 11.84IADD199 pKa = 4.3TIIDD203 pKa = 3.93TAPGEE208 pKa = 4.25FPGRR212 pKa = 11.84GMPLGVEE219 pKa = 4.64PSQQEE224 pKa = 4.02MAAMPSAVDD233 pKa = 3.41NWIKK237 pKa = 10.73CQMSTHH243 pKa = 6.08SAGHH247 pKa = 5.94YY248 pKa = 9.21MDD250 pKa = 6.15DD251 pKa = 3.74YY252 pKa = 11.48CIILPDD258 pKa = 3.92IEE260 pKa = 4.63DD261 pKa = 3.67LKK263 pKa = 11.32KK264 pKa = 10.53LGRR267 pKa = 11.84AIVRR271 pKa = 11.84QFEE274 pKa = 4.09IRR276 pKa = 11.84GIPVNKK282 pKa = 9.31KK283 pKa = 7.86KK284 pKa = 10.95CKK286 pKa = 9.78IIPLTKK292 pKa = 9.86PFRR295 pKa = 11.84WCKK298 pKa = 10.54ARR300 pKa = 11.84FTLTEE305 pKa = 3.75TGKK308 pKa = 10.45IKK310 pKa = 11.09VNGSRR315 pKa = 11.84DD316 pKa = 3.22GVIRR320 pKa = 11.84ARR322 pKa = 11.84RR323 pKa = 11.84KK324 pKa = 9.4LKK326 pKa = 10.34LFHH329 pKa = 6.95RR330 pKa = 11.84EE331 pKa = 3.17WLAGKK336 pKa = 8.25RR337 pKa = 11.84TLQEE341 pKa = 3.39VAQYY345 pKa = 8.11MNCQEE350 pKa = 4.59AYY352 pKa = 9.96YY353 pKa = 11.05KK354 pKa = 11.01NFDD357 pKa = 3.28DD358 pKa = 4.76HH359 pKa = 6.96GRR361 pKa = 11.84LLRR364 pKa = 11.84LRR366 pKa = 11.84RR367 pKa = 11.84LCYY370 pKa = 10.04AIFGGRR376 pKa = 11.84VPCSTKK382 pKa = 10.19SSKK385 pKa = 10.49PVMAPSLPP393 pKa = 3.58
MM1 pKa = 7.4TSQEE5 pKa = 3.83RR6 pKa = 11.84HH7 pKa = 4.26EE8 pKa = 4.47ARR10 pKa = 11.84YY11 pKa = 8.6QRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84AARR18 pKa = 11.84RR19 pKa = 11.84ARR21 pKa = 11.84QEE23 pKa = 3.75ARR25 pKa = 11.84CAALGSLGEE34 pKa = 4.26VFSYY38 pKa = 8.7HH39 pKa = 6.0TMFKK43 pKa = 10.6YY44 pKa = 10.39GRR46 pKa = 11.84KK47 pKa = 8.33CCNGIRR53 pKa = 11.84WKK55 pKa = 10.81QSTQNFEE62 pKa = 3.85RR63 pKa = 11.84HH64 pKa = 5.69LFSHH68 pKa = 6.08TAKK71 pKa = 9.83QRR73 pKa = 11.84RR74 pKa = 11.84LILAKK79 pKa = 9.61RR80 pKa = 11.84WRR82 pKa = 11.84PKK84 pKa = 10.35KK85 pKa = 9.19YY86 pKa = 9.97VHH88 pKa = 5.94FTVCEE93 pKa = 3.79RR94 pKa = 11.84GKK96 pKa = 10.3IRR98 pKa = 11.84GIDD101 pKa = 3.48APHH104 pKa = 5.75ITDD107 pKa = 3.64RR108 pKa = 11.84QIHH111 pKa = 5.31KK112 pKa = 10.37VISKK116 pKa = 10.21EE117 pKa = 4.0VLEE120 pKa = 4.27PLYY123 pKa = 10.31DD124 pKa = 3.52PSMIYY129 pKa = 10.77DD130 pKa = 3.68NGASRR135 pKa = 11.84IGKK138 pKa = 8.72GLHH141 pKa = 4.59WQIKK145 pKa = 9.66RR146 pKa = 11.84IKK148 pKa = 8.76QQLARR153 pKa = 11.84HH154 pKa = 4.47YY155 pKa = 9.83RR156 pKa = 11.84KK157 pKa = 9.73YY158 pKa = 10.87GRR160 pKa = 11.84AGGVLLLDD168 pKa = 4.04LKK170 pKa = 11.15KK171 pKa = 10.3FFPYY175 pKa = 10.15APHH178 pKa = 6.43SIIYY182 pKa = 9.29QRR184 pKa = 11.84HH185 pKa = 3.56QRR187 pKa = 11.84YY188 pKa = 8.88ILNPDD193 pKa = 3.54FRR195 pKa = 11.84RR196 pKa = 11.84IADD199 pKa = 4.3TIIDD203 pKa = 3.93TAPGEE208 pKa = 4.25FPGRR212 pKa = 11.84GMPLGVEE219 pKa = 4.64PSQQEE224 pKa = 4.02MAAMPSAVDD233 pKa = 3.41NWIKK237 pKa = 10.73CQMSTHH243 pKa = 6.08SAGHH247 pKa = 5.94YY248 pKa = 9.21MDD250 pKa = 6.15DD251 pKa = 3.74YY252 pKa = 11.48CIILPDD258 pKa = 3.92IEE260 pKa = 4.63DD261 pKa = 3.67LKK263 pKa = 11.32KK264 pKa = 10.53LGRR267 pKa = 11.84AIVRR271 pKa = 11.84QFEE274 pKa = 4.09IRR276 pKa = 11.84GIPVNKK282 pKa = 9.31KK283 pKa = 7.86KK284 pKa = 10.95CKK286 pKa = 9.78IIPLTKK292 pKa = 9.86PFRR295 pKa = 11.84WCKK298 pKa = 10.54ARR300 pKa = 11.84FTLTEE305 pKa = 3.75TGKK308 pKa = 10.45IKK310 pKa = 11.09VNGSRR315 pKa = 11.84DD316 pKa = 3.22GVIRR320 pKa = 11.84ARR322 pKa = 11.84RR323 pKa = 11.84KK324 pKa = 9.4LKK326 pKa = 10.34LFHH329 pKa = 6.95RR330 pKa = 11.84EE331 pKa = 3.17WLAGKK336 pKa = 8.25RR337 pKa = 11.84TLQEE341 pKa = 3.39VAQYY345 pKa = 8.11MNCQEE350 pKa = 4.59AYY352 pKa = 9.96YY353 pKa = 11.05KK354 pKa = 11.01NFDD357 pKa = 3.28DD358 pKa = 4.76HH359 pKa = 6.96GRR361 pKa = 11.84LLRR364 pKa = 11.84LRR366 pKa = 11.84RR367 pKa = 11.84LCYY370 pKa = 10.04AIFGGRR376 pKa = 11.84VPCSTKK382 pKa = 10.19SSKK385 pKa = 10.49PVMAPSLPP393 pKa = 3.58
Molecular weight: 45.93 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
16963 |
32 |
1189 |
199.6 |
22.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.939 ± 0.501 | 1.715 ± 0.138 |
6.29 ± 0.219 | 6.992 ± 0.329 |
3.142 ± 0.196 | 7.186 ± 0.249 |
1.916 ± 0.158 | 5.494 ± 0.181 |
5.995 ± 0.236 | 7.811 ± 0.267 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.847 ± 0.14 | 3.808 ± 0.159 |
3.679 ± 0.254 | 3.679 ± 0.241 |
5.241 ± 0.395 | 6.102 ± 0.386 |
6.432 ± 0.345 | 6.408 ± 0.215 |
1.462 ± 0.116 | 3.861 ± 0.161 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
