
Caenispirillum bisanense
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodospirillales; Rhodospirillaceae; Caenispirillum
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
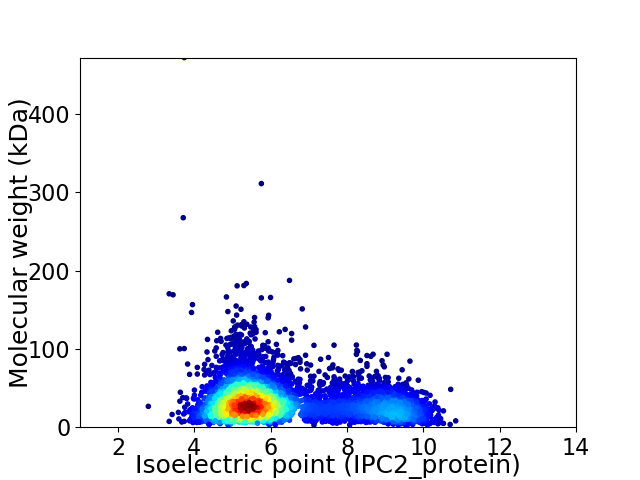
Virtual 2D-PAGE plot for 4942 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A286H2A1|A0A286H2A1_9PROT Transposase OS=Caenispirillum bisanense OX=414052 GN=SAMN05421508_1302 PE=4 SV=1
MM1 pKa = 8.04PKK3 pKa = 9.75PQPTSTRR10 pKa = 11.84TPLFDD15 pKa = 3.5TDD17 pKa = 3.96GFTSALVVDD26 pKa = 4.7AALASEE32 pKa = 4.4ISYY35 pKa = 10.93GRR37 pKa = 11.84GDD39 pKa = 4.33LDD41 pKa = 3.74AALDD45 pKa = 3.84AGGFTNVTADD55 pKa = 2.71IGLPAEE61 pKa = 4.08YY62 pKa = 9.78LVNGFFVNGNASGIVLEE79 pKa = 4.55KK80 pKa = 11.14DD81 pKa = 3.06GVLYY85 pKa = 10.81VAFAGTSVEE94 pKa = 4.34NPLPDD99 pKa = 4.39VLDD102 pKa = 3.53YY103 pKa = 10.5WQLNGQAMIDD113 pKa = 3.79EE114 pKa = 4.86YY115 pKa = 11.44YY116 pKa = 11.19GDD118 pKa = 3.78VSAMMDD124 pKa = 3.06AAMTYY129 pKa = 10.65AADD132 pKa = 4.09PEE134 pKa = 4.71SGVTQVVATGHH145 pKa = 5.89SLGGSAVDD153 pKa = 4.06GALAAYY159 pKa = 9.34DD160 pKa = 3.58AVEE163 pKa = 4.11YY164 pKa = 9.16PALRR168 pKa = 11.84GITFNSPTSGLLDD181 pKa = 4.41DD182 pKa = 5.02GRR184 pKa = 11.84EE185 pKa = 4.08LNVGYY190 pKa = 10.62EE191 pKa = 3.79NDD193 pKa = 4.7FINEE197 pKa = 4.15IIGDD201 pKa = 4.09GDD203 pKa = 3.67AGNAVHH209 pKa = 6.53NLYY212 pKa = 10.68YY213 pKa = 10.44AIQPGEE219 pKa = 4.13PMPDD223 pKa = 3.41PGSEE227 pKa = 3.68TSYY230 pKa = 10.98IALLSKK236 pKa = 10.55ALQAWTSPAHH246 pKa = 6.81DD247 pKa = 3.11IARR250 pKa = 11.84FPEE253 pKa = 4.17ALAALTEE260 pKa = 4.42SPVYY264 pKa = 9.77EE265 pKa = 4.03QVSRR269 pKa = 11.84GDD271 pKa = 3.56VVVLDD276 pKa = 3.95GSRR279 pKa = 11.84YY280 pKa = 9.35GVSLDD285 pKa = 3.84AMADD289 pKa = 3.75LSPAFDD295 pKa = 3.45AAMAGSSRR303 pKa = 11.84VGLVGQAVDD312 pKa = 4.09DD313 pKa = 4.17VLGGSAYY320 pKa = 10.53GDD322 pKa = 4.64AIDD325 pKa = 5.5GGAGDD330 pKa = 5.25DD331 pKa = 4.24VIAGLGGNDD340 pKa = 4.3LLWGGEE346 pKa = 4.08GLDD349 pKa = 3.42TFLFTSRR356 pKa = 11.84FGTDD360 pKa = 2.79VVKK363 pKa = 10.53DD364 pKa = 3.82YY365 pKa = 11.14EE366 pKa = 5.35AGDD369 pKa = 3.41ILSFQTGPAGGEE381 pKa = 4.05YY382 pKa = 10.24LASLAALQAWTEE394 pKa = 4.13STNQIRR400 pKa = 11.84TSFTDD405 pKa = 2.96AGLVIAFAGGKK416 pKa = 10.1DD417 pKa = 3.27KK418 pKa = 10.85VTLEE422 pKa = 4.51GVGSWEE428 pKa = 4.46LMAA431 pKa = 6.62
MM1 pKa = 8.04PKK3 pKa = 9.75PQPTSTRR10 pKa = 11.84TPLFDD15 pKa = 3.5TDD17 pKa = 3.96GFTSALVVDD26 pKa = 4.7AALASEE32 pKa = 4.4ISYY35 pKa = 10.93GRR37 pKa = 11.84GDD39 pKa = 4.33LDD41 pKa = 3.74AALDD45 pKa = 3.84AGGFTNVTADD55 pKa = 2.71IGLPAEE61 pKa = 4.08YY62 pKa = 9.78LVNGFFVNGNASGIVLEE79 pKa = 4.55KK80 pKa = 11.14DD81 pKa = 3.06GVLYY85 pKa = 10.81VAFAGTSVEE94 pKa = 4.34NPLPDD99 pKa = 4.39VLDD102 pKa = 3.53YY103 pKa = 10.5WQLNGQAMIDD113 pKa = 3.79EE114 pKa = 4.86YY115 pKa = 11.44YY116 pKa = 11.19GDD118 pKa = 3.78VSAMMDD124 pKa = 3.06AAMTYY129 pKa = 10.65AADD132 pKa = 4.09PEE134 pKa = 4.71SGVTQVVATGHH145 pKa = 5.89SLGGSAVDD153 pKa = 4.06GALAAYY159 pKa = 9.34DD160 pKa = 3.58AVEE163 pKa = 4.11YY164 pKa = 9.16PALRR168 pKa = 11.84GITFNSPTSGLLDD181 pKa = 4.41DD182 pKa = 5.02GRR184 pKa = 11.84EE185 pKa = 4.08LNVGYY190 pKa = 10.62EE191 pKa = 3.79NDD193 pKa = 4.7FINEE197 pKa = 4.15IIGDD201 pKa = 4.09GDD203 pKa = 3.67AGNAVHH209 pKa = 6.53NLYY212 pKa = 10.68YY213 pKa = 10.44AIQPGEE219 pKa = 4.13PMPDD223 pKa = 3.41PGSEE227 pKa = 3.68TSYY230 pKa = 10.98IALLSKK236 pKa = 10.55ALQAWTSPAHH246 pKa = 6.81DD247 pKa = 3.11IARR250 pKa = 11.84FPEE253 pKa = 4.17ALAALTEE260 pKa = 4.42SPVYY264 pKa = 9.77EE265 pKa = 4.03QVSRR269 pKa = 11.84GDD271 pKa = 3.56VVVLDD276 pKa = 3.95GSRR279 pKa = 11.84YY280 pKa = 9.35GVSLDD285 pKa = 3.84AMADD289 pKa = 3.75LSPAFDD295 pKa = 3.45AAMAGSSRR303 pKa = 11.84VGLVGQAVDD312 pKa = 4.09DD313 pKa = 4.17VLGGSAYY320 pKa = 10.53GDD322 pKa = 4.64AIDD325 pKa = 5.5GGAGDD330 pKa = 5.25DD331 pKa = 4.24VIAGLGGNDD340 pKa = 4.3LLWGGEE346 pKa = 4.08GLDD349 pKa = 3.42TFLFTSRR356 pKa = 11.84FGTDD360 pKa = 2.79VVKK363 pKa = 10.53DD364 pKa = 3.82YY365 pKa = 11.14EE366 pKa = 5.35AGDD369 pKa = 3.41ILSFQTGPAGGEE381 pKa = 4.05YY382 pKa = 10.24LASLAALQAWTEE394 pKa = 4.13STNQIRR400 pKa = 11.84TSFTDD405 pKa = 2.96AGLVIAFAGGKK416 pKa = 10.1DD417 pKa = 3.27KK418 pKa = 10.85VTLEE422 pKa = 4.51GVGSWEE428 pKa = 4.46LMAA431 pKa = 6.62
Molecular weight: 44.6 kDa
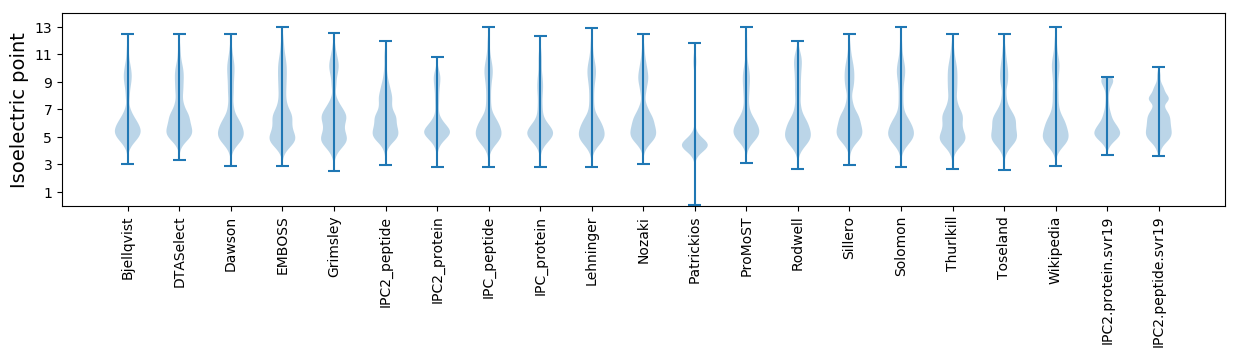
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A286H0I6|A0A286H0I6_9PROT TRAP-type mannitol/chloroaromatic compound transport system small permease component OS=Caenispirillum bisanense OX=414052 GN=SAMN05421508_11450 PE=4 SV=1
MM1 pKa = 7.06NLWLRR6 pKa = 11.84LLRR9 pKa = 11.84IIVGALLGRR18 pKa = 11.84RR19 pKa = 11.84LGMFDD24 pKa = 3.02TSEE27 pKa = 3.6LRR29 pKa = 11.84FRR31 pKa = 11.84VWPHH35 pKa = 6.72DD36 pKa = 3.91LDD38 pKa = 5.79LNIHH42 pKa = 5.81MNNARR47 pKa = 11.84YY48 pKa = 9.47LSLMDD53 pKa = 5.06LGRR56 pKa = 11.84ADD58 pKa = 4.95LIIRR62 pKa = 11.84TGLHH66 pKa = 4.22TVMLRR71 pKa = 11.84EE72 pKa = 4.22GWQAVIGGTTVRR84 pKa = 11.84FRR86 pKa = 11.84RR87 pKa = 11.84PLKK90 pKa = 10.14PFQACRR96 pKa = 11.84LTTRR100 pKa = 11.84LLCWDD105 pKa = 4.5DD106 pKa = 2.71RR107 pKa = 11.84WLYY110 pKa = 10.06IEE112 pKa = 5.32HH113 pKa = 7.73RR114 pKa = 11.84ITTADD119 pKa = 3.26GTLACAALVKK129 pKa = 10.61AVFIRR134 pKa = 11.84KK135 pKa = 9.24GSRR138 pKa = 11.84VAPAEE143 pKa = 3.94TVARR147 pKa = 11.84IGVTTPSPAAPDD159 pKa = 3.5WVKK162 pKa = 10.57VPEE165 pKa = 4.32VVPEE169 pKa = 3.88
MM1 pKa = 7.06NLWLRR6 pKa = 11.84LLRR9 pKa = 11.84IIVGALLGRR18 pKa = 11.84RR19 pKa = 11.84LGMFDD24 pKa = 3.02TSEE27 pKa = 3.6LRR29 pKa = 11.84FRR31 pKa = 11.84VWPHH35 pKa = 6.72DD36 pKa = 3.91LDD38 pKa = 5.79LNIHH42 pKa = 5.81MNNARR47 pKa = 11.84YY48 pKa = 9.47LSLMDD53 pKa = 5.06LGRR56 pKa = 11.84ADD58 pKa = 4.95LIIRR62 pKa = 11.84TGLHH66 pKa = 4.22TVMLRR71 pKa = 11.84EE72 pKa = 4.22GWQAVIGGTTVRR84 pKa = 11.84FRR86 pKa = 11.84RR87 pKa = 11.84PLKK90 pKa = 10.14PFQACRR96 pKa = 11.84LTTRR100 pKa = 11.84LLCWDD105 pKa = 4.5DD106 pKa = 2.71RR107 pKa = 11.84WLYY110 pKa = 10.06IEE112 pKa = 5.32HH113 pKa = 7.73RR114 pKa = 11.84ITTADD119 pKa = 3.26GTLACAALVKK129 pKa = 10.61AVFIRR134 pKa = 11.84KK135 pKa = 9.24GSRR138 pKa = 11.84VAPAEE143 pKa = 3.94TVARR147 pKa = 11.84IGVTTPSPAAPDD159 pKa = 3.5WVKK162 pKa = 10.57VPEE165 pKa = 4.32VVPEE169 pKa = 3.88
Molecular weight: 19.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1577073 |
24 |
4564 |
319.1 |
34.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.921 ± 0.059 | 0.862 ± 0.012 |
6.085 ± 0.03 | 5.73 ± 0.033 |
3.314 ± 0.021 | 8.832 ± 0.033 |
2.041 ± 0.017 | 4.035 ± 0.025 |
2.677 ± 0.027 | 10.436 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.411 ± 0.018 | 1.993 ± 0.018 |
5.599 ± 0.03 | 2.86 ± 0.019 |
7.663 ± 0.046 | 4.282 ± 0.024 |
5.493 ± 0.035 | 8.355 ± 0.03 |
1.371 ± 0.014 | 2.04 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
