
Raphanus sativus cryptic virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; unclassified Partitiviridae
Average proteome isoelectric point is 5.84
Get precalculated fractions of proteins
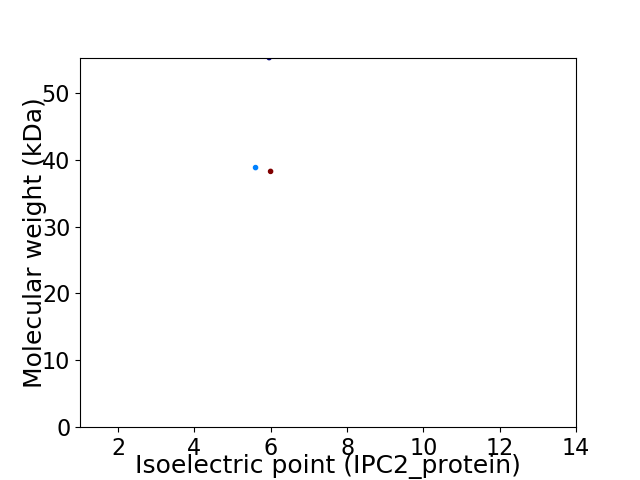
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q38L13|Q38L13_9VIRU Uncharacterized protein OS=Raphanus sativus cryptic virus 2 OX=351495 PE=4 SV=1
MM1 pKa = 7.73DD2 pKa = 4.61TAQTDD7 pKa = 3.26ADD9 pKa = 3.72KK10 pKa = 11.32AAGKK14 pKa = 10.15RR15 pKa = 11.84GATVPPEE22 pKa = 4.22GEE24 pKa = 3.91PAAKK28 pKa = 9.71TMRR31 pKa = 11.84FSDD34 pKa = 4.67AIVPAGPSKK43 pKa = 10.73DD44 pKa = 3.26ASTVATAALPKK55 pKa = 10.55DD56 pKa = 4.45LIQWPIEE63 pKa = 3.92RR64 pKa = 11.84LATQVNQVFLDD75 pKa = 3.62TTIQNGEE82 pKa = 3.98GGYY85 pKa = 9.54EE86 pKa = 4.06VPDD89 pKa = 3.96DD90 pKa = 4.16QIVPTLRR97 pKa = 11.84VYY99 pKa = 10.28EE100 pKa = 4.15INVTNLRR107 pKa = 11.84NAFRR111 pKa = 11.84SVLYY115 pKa = 9.89TILRR119 pKa = 11.84AKK121 pKa = 10.37FIEE124 pKa = 4.56RR125 pKa = 11.84GDD127 pKa = 3.84RR128 pKa = 11.84TVTAATTLATNVSNLTADD146 pKa = 3.13ACMAALYY153 pKa = 10.44AKK155 pKa = 10.12LRR157 pKa = 11.84SLHH160 pKa = 5.38RR161 pKa = 11.84QIYY164 pKa = 4.99THH166 pKa = 7.14KK167 pKa = 10.8GRR169 pKa = 11.84FTDD172 pKa = 3.41QPTYY176 pKa = 10.21TKK178 pKa = 10.5DD179 pKa = 3.3VEE181 pKa = 4.5LPLPFAVAIDD191 pKa = 4.49GIGMFRR197 pKa = 11.84TSAMSTRR204 pKa = 11.84FNVVPVYY211 pKa = 10.49PEE213 pKa = 3.59NTKK216 pKa = 10.89NEE218 pKa = 4.06GRR220 pKa = 11.84SHH222 pKa = 7.47DD223 pKa = 4.14SINFLEE229 pKa = 4.35YY230 pKa = 10.38KK231 pKa = 10.53SYY233 pKa = 10.87LAYY236 pKa = 10.02FAEE239 pKa = 4.49IGIPTRR245 pKa = 11.84TIDD248 pKa = 3.75TRR250 pKa = 11.84VTPGNAWWTYY260 pKa = 6.93RR261 pKa = 11.84TSYY264 pKa = 11.17DD265 pKa = 3.19GDD267 pKa = 3.87VYY269 pKa = 10.97DD270 pKa = 5.54LKK272 pKa = 11.24INFPPLHH279 pKa = 6.49FNDD282 pKa = 4.17HH283 pKa = 6.4LANLAIMFLGNSGADD298 pKa = 3.33RR299 pKa = 11.84NNASIITTKK308 pKa = 10.52TDD310 pKa = 3.18DD311 pKa = 3.92QDD313 pKa = 3.91YY314 pKa = 9.56GWYY317 pKa = 10.37VKK319 pKa = 10.25DD320 pKa = 3.34IRR322 pKa = 11.84EE323 pKa = 4.42GEE325 pKa = 4.08QARR328 pKa = 11.84AFAALCQWDD337 pKa = 3.63PRR339 pKa = 11.84YY340 pKa = 9.87WNEE343 pKa = 3.85KK344 pKa = 9.77NNVV347 pKa = 3.21
MM1 pKa = 7.73DD2 pKa = 4.61TAQTDD7 pKa = 3.26ADD9 pKa = 3.72KK10 pKa = 11.32AAGKK14 pKa = 10.15RR15 pKa = 11.84GATVPPEE22 pKa = 4.22GEE24 pKa = 3.91PAAKK28 pKa = 9.71TMRR31 pKa = 11.84FSDD34 pKa = 4.67AIVPAGPSKK43 pKa = 10.73DD44 pKa = 3.26ASTVATAALPKK55 pKa = 10.55DD56 pKa = 4.45LIQWPIEE63 pKa = 3.92RR64 pKa = 11.84LATQVNQVFLDD75 pKa = 3.62TTIQNGEE82 pKa = 3.98GGYY85 pKa = 9.54EE86 pKa = 4.06VPDD89 pKa = 3.96DD90 pKa = 4.16QIVPTLRR97 pKa = 11.84VYY99 pKa = 10.28EE100 pKa = 4.15INVTNLRR107 pKa = 11.84NAFRR111 pKa = 11.84SVLYY115 pKa = 9.89TILRR119 pKa = 11.84AKK121 pKa = 10.37FIEE124 pKa = 4.56RR125 pKa = 11.84GDD127 pKa = 3.84RR128 pKa = 11.84TVTAATTLATNVSNLTADD146 pKa = 3.13ACMAALYY153 pKa = 10.44AKK155 pKa = 10.12LRR157 pKa = 11.84SLHH160 pKa = 5.38RR161 pKa = 11.84QIYY164 pKa = 4.99THH166 pKa = 7.14KK167 pKa = 10.8GRR169 pKa = 11.84FTDD172 pKa = 3.41QPTYY176 pKa = 10.21TKK178 pKa = 10.5DD179 pKa = 3.3VEE181 pKa = 4.5LPLPFAVAIDD191 pKa = 4.49GIGMFRR197 pKa = 11.84TSAMSTRR204 pKa = 11.84FNVVPVYY211 pKa = 10.49PEE213 pKa = 3.59NTKK216 pKa = 10.89NEE218 pKa = 4.06GRR220 pKa = 11.84SHH222 pKa = 7.47DD223 pKa = 4.14SINFLEE229 pKa = 4.35YY230 pKa = 10.38KK231 pKa = 10.53SYY233 pKa = 10.87LAYY236 pKa = 10.02FAEE239 pKa = 4.49IGIPTRR245 pKa = 11.84TIDD248 pKa = 3.75TRR250 pKa = 11.84VTPGNAWWTYY260 pKa = 6.93RR261 pKa = 11.84TSYY264 pKa = 11.17DD265 pKa = 3.19GDD267 pKa = 3.87VYY269 pKa = 10.97DD270 pKa = 5.54LKK272 pKa = 11.24INFPPLHH279 pKa = 6.49FNDD282 pKa = 4.17HH283 pKa = 6.4LANLAIMFLGNSGADD298 pKa = 3.33RR299 pKa = 11.84NNASIITTKK308 pKa = 10.52TDD310 pKa = 3.18DD311 pKa = 3.92QDD313 pKa = 3.91YY314 pKa = 9.56GWYY317 pKa = 10.37VKK319 pKa = 10.25DD320 pKa = 3.34IRR322 pKa = 11.84EE323 pKa = 4.42GEE325 pKa = 4.08QARR328 pKa = 11.84AFAALCQWDD337 pKa = 3.63PRR339 pKa = 11.84YY340 pKa = 9.87WNEE343 pKa = 3.85KK344 pKa = 9.77NNVV347 pKa = 3.21
Molecular weight: 38.81 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q38L14|Q38L14_9VIRU Putative RNA dependent RNA polymerse OS=Raphanus sativus cryptic virus 2 OX=351495 PE=4 SV=1
MM1 pKa = 7.5AFEE4 pKa = 5.06SKK6 pKa = 9.47TSASEE11 pKa = 4.01STVNKK16 pKa = 9.88DD17 pKa = 3.04AAPAPSSASVKK28 pKa = 10.77SPVKK32 pKa = 10.01PSSSGLGRR40 pKa = 11.84AKK42 pKa = 10.07TEE44 pKa = 4.31TPHH47 pKa = 7.27LPASNYY53 pKa = 7.79EE54 pKa = 3.83IPRR57 pKa = 11.84YY58 pKa = 8.2TRR60 pKa = 11.84DD61 pKa = 3.38TQPVVGNKK69 pKa = 9.84NYY71 pKa = 10.17EE72 pKa = 3.66FSLRR76 pKa = 11.84MDD78 pKa = 3.2GRR80 pKa = 11.84LGHH83 pKa = 6.43SMDD86 pKa = 3.36RR87 pKa = 11.84SKK89 pKa = 9.87TARR92 pKa = 11.84PEE94 pKa = 4.27VNCSLNRR101 pKa = 11.84ANFHH105 pKa = 6.1EE106 pKa = 5.07GVLKK110 pKa = 10.34VLRR113 pKa = 11.84QILSEE118 pKa = 4.04MARR121 pKa = 11.84LKK123 pKa = 10.58QDD125 pKa = 3.37LSDD128 pKa = 4.67AEE130 pKa = 3.85IQEE133 pKa = 4.41RR134 pKa = 11.84VEE136 pKa = 4.56SVSLTLAYY144 pKa = 9.94GVATMAYY151 pKa = 9.68LKK153 pKa = 10.66LRR155 pKa = 11.84AINLFRR161 pKa = 11.84PNEE164 pKa = 3.8ASKK167 pKa = 10.72FLTKK171 pKa = 10.4PKK173 pKa = 10.54VPDD176 pKa = 3.32HH177 pKa = 6.73FEE179 pKa = 3.86IPTPFAFAISQLGVVEE195 pKa = 4.41VSSLSRR201 pKa = 11.84RR202 pKa = 11.84MICYY206 pKa = 7.61PTADD210 pKa = 4.1LADD213 pKa = 4.09ASNHH217 pKa = 5.19LVGNKK222 pKa = 9.82RR223 pKa = 11.84NWSQTAYY230 pKa = 10.68AEE232 pKa = 4.09AVRR235 pKa = 11.84YY236 pKa = 9.31AKK238 pKa = 10.55YY239 pKa = 10.71LGMSFSTVDD248 pKa = 3.7LDD250 pKa = 4.15IKK252 pKa = 11.02VGSSWWLFKK261 pKa = 10.72PDD263 pKa = 3.45VTDD266 pKa = 3.61GLLSIRR272 pKa = 11.84CPLPEE277 pKa = 5.58DD278 pKa = 3.91NYY280 pKa = 9.19TLAGATVFMLFYY292 pKa = 10.11HH293 pKa = 7.28DD294 pKa = 4.58VGSDD298 pKa = 3.4PAIDD302 pKa = 4.43LFNIDD307 pKa = 3.87SLGNDD312 pKa = 4.03DD313 pKa = 4.2YY314 pKa = 12.03GSFIRR319 pKa = 11.84NPRR322 pKa = 11.84DD323 pKa = 3.03GFNASSYY330 pKa = 10.17YY331 pKa = 10.64AISSEE336 pKa = 4.25GTDD339 pKa = 3.78EE340 pKa = 4.08MWKK343 pKa = 10.62SSAA346 pKa = 3.97
MM1 pKa = 7.5AFEE4 pKa = 5.06SKK6 pKa = 9.47TSASEE11 pKa = 4.01STVNKK16 pKa = 9.88DD17 pKa = 3.04AAPAPSSASVKK28 pKa = 10.77SPVKK32 pKa = 10.01PSSSGLGRR40 pKa = 11.84AKK42 pKa = 10.07TEE44 pKa = 4.31TPHH47 pKa = 7.27LPASNYY53 pKa = 7.79EE54 pKa = 3.83IPRR57 pKa = 11.84YY58 pKa = 8.2TRR60 pKa = 11.84DD61 pKa = 3.38TQPVVGNKK69 pKa = 9.84NYY71 pKa = 10.17EE72 pKa = 3.66FSLRR76 pKa = 11.84MDD78 pKa = 3.2GRR80 pKa = 11.84LGHH83 pKa = 6.43SMDD86 pKa = 3.36RR87 pKa = 11.84SKK89 pKa = 9.87TARR92 pKa = 11.84PEE94 pKa = 4.27VNCSLNRR101 pKa = 11.84ANFHH105 pKa = 6.1EE106 pKa = 5.07GVLKK110 pKa = 10.34VLRR113 pKa = 11.84QILSEE118 pKa = 4.04MARR121 pKa = 11.84LKK123 pKa = 10.58QDD125 pKa = 3.37LSDD128 pKa = 4.67AEE130 pKa = 3.85IQEE133 pKa = 4.41RR134 pKa = 11.84VEE136 pKa = 4.56SVSLTLAYY144 pKa = 9.94GVATMAYY151 pKa = 9.68LKK153 pKa = 10.66LRR155 pKa = 11.84AINLFRR161 pKa = 11.84PNEE164 pKa = 3.8ASKK167 pKa = 10.72FLTKK171 pKa = 10.4PKK173 pKa = 10.54VPDD176 pKa = 3.32HH177 pKa = 6.73FEE179 pKa = 3.86IPTPFAFAISQLGVVEE195 pKa = 4.41VSSLSRR201 pKa = 11.84RR202 pKa = 11.84MICYY206 pKa = 7.61PTADD210 pKa = 4.1LADD213 pKa = 4.09ASNHH217 pKa = 5.19LVGNKK222 pKa = 9.82RR223 pKa = 11.84NWSQTAYY230 pKa = 10.68AEE232 pKa = 4.09AVRR235 pKa = 11.84YY236 pKa = 9.31AKK238 pKa = 10.55YY239 pKa = 10.71LGMSFSTVDD248 pKa = 3.7LDD250 pKa = 4.15IKK252 pKa = 11.02VGSSWWLFKK261 pKa = 10.72PDD263 pKa = 3.45VTDD266 pKa = 3.61GLLSIRR272 pKa = 11.84CPLPEE277 pKa = 5.58DD278 pKa = 3.91NYY280 pKa = 9.19TLAGATVFMLFYY292 pKa = 10.11HH293 pKa = 7.28DD294 pKa = 4.58VGSDD298 pKa = 3.4PAIDD302 pKa = 4.43LFNIDD307 pKa = 3.87SLGNDD312 pKa = 4.03DD313 pKa = 4.2YY314 pKa = 12.03GSFIRR319 pKa = 11.84NPRR322 pKa = 11.84DD323 pKa = 3.03GFNASSYY330 pKa = 10.17YY331 pKa = 10.64AISSEE336 pKa = 4.25GTDD339 pKa = 3.78EE340 pKa = 4.08MWKK343 pKa = 10.62SSAA346 pKa = 3.97
Molecular weight: 38.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1170 |
346 |
477 |
390.0 |
44.1 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.632 ± 1.116 | 0.769 ± 0.069 |
6.325 ± 0.496 | 5.556 ± 0.6 |
5.128 ± 0.675 | 5.043 ± 0.22 |
1.88 ± 0.219 | 5.556 ± 0.665 |
5.043 ± 0.274 | 8.034 ± 0.506 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.88 ± 0.256 | 4.701 ± 0.613 |
4.957 ± 0.474 | 3.162 ± 0.559 |
5.983 ± 0.128 | 7.863 ± 1.553 |
6.581 ± 1.162 | 6.496 ± 0.219 |
1.624 ± 0.165 | 4.786 ± 0.335 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
