
Odonata-associated circular virus-7
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
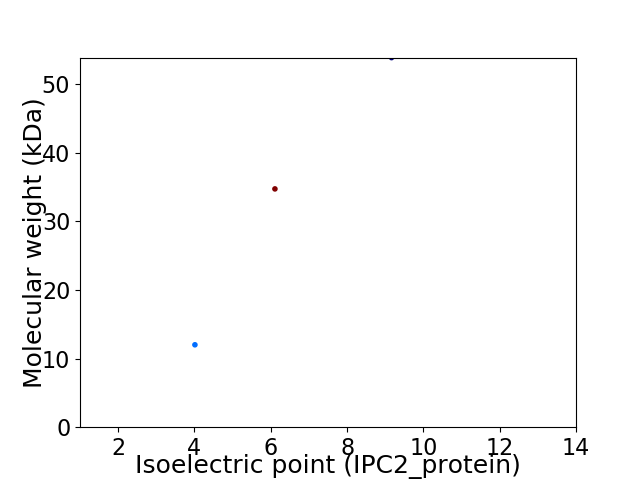
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
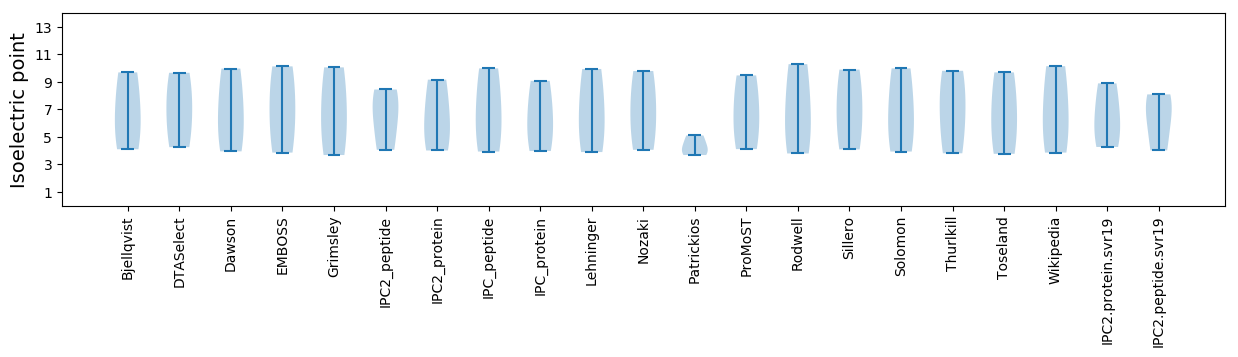
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UH28|A0A0B4UH28_9VIRU Putative capsid protein OS=Odonata-associated circular virus-7 OX=1592127 PE=4 SV=1
MM1 pKa = 7.45FNLDD5 pKa = 3.55GAKK8 pKa = 10.17SCDD11 pKa = 3.69FQDD14 pKa = 4.09PCWCEE19 pKa = 3.74YY20 pKa = 10.4IRR22 pKa = 11.84TEE24 pKa = 3.93GMGYY28 pKa = 10.21IMDD31 pKa = 4.53IDD33 pKa = 4.07GADD36 pKa = 3.07ITEE39 pKa = 4.22FQPDD43 pKa = 3.65EE44 pKa = 4.99FIDD47 pKa = 4.29LGDD50 pKa = 3.88SRR52 pKa = 11.84DD53 pKa = 3.85DD54 pKa = 3.46EE55 pKa = 4.92TIVISSDD62 pKa = 3.7SEE64 pKa = 4.07DD65 pKa = 3.54DD66 pKa = 3.87KK67 pKa = 12.01EE68 pKa = 4.99DD69 pKa = 3.84IEE71 pKa = 4.21WLEE74 pKa = 4.43EE75 pKa = 3.51IRR77 pKa = 11.84KK78 pKa = 7.97RR79 pKa = 11.84TRR81 pKa = 11.84LMKK84 pKa = 10.48RR85 pKa = 11.84EE86 pKa = 3.88IVKK89 pKa = 10.07AACEE93 pKa = 3.82QLAEE97 pKa = 4.01EE98 pKa = 5.49LINKK102 pKa = 7.94NN103 pKa = 3.06
MM1 pKa = 7.45FNLDD5 pKa = 3.55GAKK8 pKa = 10.17SCDD11 pKa = 3.69FQDD14 pKa = 4.09PCWCEE19 pKa = 3.74YY20 pKa = 10.4IRR22 pKa = 11.84TEE24 pKa = 3.93GMGYY28 pKa = 10.21IMDD31 pKa = 4.53IDD33 pKa = 4.07GADD36 pKa = 3.07ITEE39 pKa = 4.22FQPDD43 pKa = 3.65EE44 pKa = 4.99FIDD47 pKa = 4.29LGDD50 pKa = 3.88SRR52 pKa = 11.84DD53 pKa = 3.85DD54 pKa = 3.46EE55 pKa = 4.92TIVISSDD62 pKa = 3.7SEE64 pKa = 4.07DD65 pKa = 3.54DD66 pKa = 3.87KK67 pKa = 12.01EE68 pKa = 4.99DD69 pKa = 3.84IEE71 pKa = 4.21WLEE74 pKa = 4.43EE75 pKa = 3.51IRR77 pKa = 11.84KK78 pKa = 7.97RR79 pKa = 11.84TRR81 pKa = 11.84LMKK84 pKa = 10.48RR85 pKa = 11.84EE86 pKa = 3.88IVKK89 pKa = 10.07AACEE93 pKa = 3.82QLAEE97 pKa = 4.01EE98 pKa = 5.49LINKK102 pKa = 7.94NN103 pKa = 3.06
Molecular weight: 12.01 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UHZ3|A0A0B4UHZ3_9VIRU Replication-associated protein OS=Odonata-associated circular virus-7 OX=1592127 PE=3 SV=1
MM1 pKa = 7.32AAVAGRR7 pKa = 11.84FALRR11 pKa = 11.84SFARR15 pKa = 11.84YY16 pKa = 8.51FPQAMRR22 pKa = 11.84YY23 pKa = 6.4MARR26 pKa = 11.84NPGQGGRR33 pKa = 11.84LLRR36 pKa = 11.84SSAKK40 pKa = 9.17YY41 pKa = 10.15ARR43 pKa = 11.84AAAPVAGLAANAVRR57 pKa = 11.84YY58 pKa = 9.45VSKK61 pKa = 10.68KK62 pKa = 9.34SKK64 pKa = 10.38QYY66 pKa = 11.27LYY68 pKa = 11.19LKK70 pKa = 9.86NLKK73 pKa = 10.04KK74 pKa = 10.7KK75 pKa = 9.95IVSSRR80 pKa = 11.84GKK82 pKa = 10.14KK83 pKa = 8.69NQPSVFKK90 pKa = 10.43KK91 pKa = 10.44RR92 pKa = 11.84RR93 pKa = 11.84ITTQGVSGKK102 pKa = 10.02RR103 pKa = 11.84LEE105 pKa = 4.23LSRR108 pKa = 11.84KK109 pKa = 9.15YY110 pKa = 10.73VDD112 pKa = 3.64PFSFYY117 pKa = 10.24NRR119 pKa = 11.84KK120 pKa = 9.29RR121 pKa = 11.84VVKK124 pKa = 10.07IMKK127 pKa = 9.93VYY129 pKa = 9.04GTEE132 pKa = 3.99QTDD135 pKa = 3.58TCCYY139 pKa = 10.05VEE141 pKa = 4.34AAVTEE146 pKa = 4.11PRR148 pKa = 11.84AWILNMWKK156 pKa = 10.49ALVRR160 pKa = 11.84KK161 pKa = 9.76LSQKK165 pKa = 10.82AGLQIYY171 pKa = 9.23SCNEE175 pKa = 4.14AISMKK180 pKa = 9.32TLGVGGVDD188 pKa = 4.16LEE190 pKa = 4.52MVLYY194 pKa = 8.21TQNQANVSAPSITVSFADD212 pKa = 3.87GTTVTSLAEE221 pKa = 4.63AIAQPFYY228 pKa = 10.79SYY230 pKa = 11.6SSGSTNAGVGSVLSLVEE247 pKa = 4.07PNYY250 pKa = 10.83LVLQRR255 pKa = 11.84KK256 pKa = 9.26SSVVNFTYY264 pKa = 10.55QIVNLQLDD272 pKa = 3.85NVLVEE277 pKa = 5.54GKK279 pKa = 9.59LCCIMKK285 pKa = 9.85LQNQNQSNTGGTDD298 pKa = 3.31SEE300 pKa = 4.77DD301 pKa = 3.74VSNHH305 pKa = 5.59PLVGKK310 pKa = 10.17LYY312 pKa = 10.23YY313 pKa = 10.79FNGLPGTKK321 pKa = 9.76VPNQDD326 pKa = 3.51PGSLEE331 pKa = 3.86NNIFRR336 pKa = 11.84RR337 pKa = 11.84FPADD341 pKa = 3.21RR342 pKa = 11.84HH343 pKa = 5.83LKK345 pKa = 9.43ILKK348 pKa = 9.54DD349 pKa = 3.54ANLEE353 pKa = 4.02QEE355 pKa = 4.2LRR357 pKa = 11.84EE358 pKa = 4.47PISPISFWNCIGCTNFKK375 pKa = 9.97LQPGEE380 pKa = 3.69IRR382 pKa = 11.84TFTFWDD388 pKa = 3.64QVKK391 pKa = 9.92SQPFLKK397 pKa = 10.34YY398 pKa = 9.84LKK400 pKa = 10.22KK401 pKa = 10.52FRR403 pKa = 11.84LEE405 pKa = 4.1YY406 pKa = 9.33GTNAPAQLQDD416 pKa = 3.55QYY418 pKa = 12.0NLNYY422 pKa = 10.22CPLKK426 pKa = 10.12FAMVGLEE433 pKa = 3.91EE434 pKa = 4.54QIQSSSSNNLTIGYY448 pKa = 9.0EE449 pKa = 3.93IEE451 pKa = 4.19SRR453 pKa = 11.84CASMFKK459 pKa = 10.15EE460 pKa = 4.11LRR462 pKa = 11.84PNFCRR467 pKa = 11.84TEE469 pKa = 4.14TEE471 pKa = 4.32TKK473 pKa = 9.46QFPVIVV479 pKa = 3.74
MM1 pKa = 7.32AAVAGRR7 pKa = 11.84FALRR11 pKa = 11.84SFARR15 pKa = 11.84YY16 pKa = 8.51FPQAMRR22 pKa = 11.84YY23 pKa = 6.4MARR26 pKa = 11.84NPGQGGRR33 pKa = 11.84LLRR36 pKa = 11.84SSAKK40 pKa = 9.17YY41 pKa = 10.15ARR43 pKa = 11.84AAAPVAGLAANAVRR57 pKa = 11.84YY58 pKa = 9.45VSKK61 pKa = 10.68KK62 pKa = 9.34SKK64 pKa = 10.38QYY66 pKa = 11.27LYY68 pKa = 11.19LKK70 pKa = 9.86NLKK73 pKa = 10.04KK74 pKa = 10.7KK75 pKa = 9.95IVSSRR80 pKa = 11.84GKK82 pKa = 10.14KK83 pKa = 8.69NQPSVFKK90 pKa = 10.43KK91 pKa = 10.44RR92 pKa = 11.84RR93 pKa = 11.84ITTQGVSGKK102 pKa = 10.02RR103 pKa = 11.84LEE105 pKa = 4.23LSRR108 pKa = 11.84KK109 pKa = 9.15YY110 pKa = 10.73VDD112 pKa = 3.64PFSFYY117 pKa = 10.24NRR119 pKa = 11.84KK120 pKa = 9.29RR121 pKa = 11.84VVKK124 pKa = 10.07IMKK127 pKa = 9.93VYY129 pKa = 9.04GTEE132 pKa = 3.99QTDD135 pKa = 3.58TCCYY139 pKa = 10.05VEE141 pKa = 4.34AAVTEE146 pKa = 4.11PRR148 pKa = 11.84AWILNMWKK156 pKa = 10.49ALVRR160 pKa = 11.84KK161 pKa = 9.76LSQKK165 pKa = 10.82AGLQIYY171 pKa = 9.23SCNEE175 pKa = 4.14AISMKK180 pKa = 9.32TLGVGGVDD188 pKa = 4.16LEE190 pKa = 4.52MVLYY194 pKa = 8.21TQNQANVSAPSITVSFADD212 pKa = 3.87GTTVTSLAEE221 pKa = 4.63AIAQPFYY228 pKa = 10.79SYY230 pKa = 11.6SSGSTNAGVGSVLSLVEE247 pKa = 4.07PNYY250 pKa = 10.83LVLQRR255 pKa = 11.84KK256 pKa = 9.26SSVVNFTYY264 pKa = 10.55QIVNLQLDD272 pKa = 3.85NVLVEE277 pKa = 5.54GKK279 pKa = 9.59LCCIMKK285 pKa = 9.85LQNQNQSNTGGTDD298 pKa = 3.31SEE300 pKa = 4.77DD301 pKa = 3.74VSNHH305 pKa = 5.59PLVGKK310 pKa = 10.17LYY312 pKa = 10.23YY313 pKa = 10.79FNGLPGTKK321 pKa = 9.76VPNQDD326 pKa = 3.51PGSLEE331 pKa = 3.86NNIFRR336 pKa = 11.84RR337 pKa = 11.84FPADD341 pKa = 3.21RR342 pKa = 11.84HH343 pKa = 5.83LKK345 pKa = 9.43ILKK348 pKa = 9.54DD349 pKa = 3.54ANLEE353 pKa = 4.02QEE355 pKa = 4.2LRR357 pKa = 11.84EE358 pKa = 4.47PISPISFWNCIGCTNFKK375 pKa = 9.97LQPGEE380 pKa = 3.69IRR382 pKa = 11.84TFTFWDD388 pKa = 3.64QVKK391 pKa = 9.92SQPFLKK397 pKa = 10.34YY398 pKa = 9.84LKK400 pKa = 10.22KK401 pKa = 10.52FRR403 pKa = 11.84LEE405 pKa = 4.1YY406 pKa = 9.33GTNAPAQLQDD416 pKa = 3.55QYY418 pKa = 12.0NLNYY422 pKa = 10.22CPLKK426 pKa = 10.12FAMVGLEE433 pKa = 3.91EE434 pKa = 4.54QIQSSSSNNLTIGYY448 pKa = 9.0EE449 pKa = 3.93IEE451 pKa = 4.19SRR453 pKa = 11.84CASMFKK459 pKa = 10.15EE460 pKa = 4.11LRR462 pKa = 11.84PNFCRR467 pKa = 11.84TEE469 pKa = 4.14TEE471 pKa = 4.32TKK473 pKa = 9.46QFPVIVV479 pKa = 3.74
Molecular weight: 53.85 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
878 |
103 |
479 |
292.7 |
33.52 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.036 ± 0.936 | 2.506 ± 0.407 |
4.897 ± 2.67 | 7.062 ± 2.046 |
5.353 ± 0.818 | 5.695 ± 0.297 |
1.822 ± 1.345 | 5.695 ± 1.475 |
6.72 ± 0.588 | 7.745 ± 1.072 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.05 ± 0.508 | 5.353 ± 0.905 |
4.556 ± 0.701 | 4.784 ± 0.816 |
6.264 ± 0.384 | 6.72 ± 1.057 |
5.467 ± 0.527 | 5.923 ± 1.351 |
1.253 ± 0.318 | 4.1 ± 0.656 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
