
Herrania umbratica
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Byttnerioideae; Herrania
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
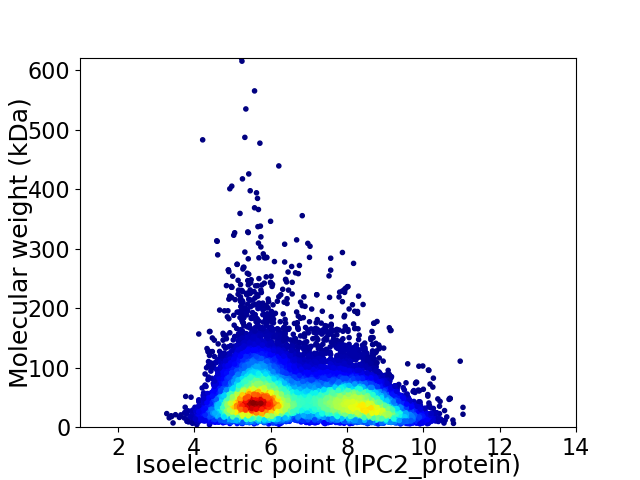
Virtual 2D-PAGE plot for 23848 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6J1BQI0|A0A6J1BQI0_9ROSI uncharacterized protein LOC110428928 OS=Herrania umbratica OX=108875 GN=LOC110428928 PE=3 SV=1
HHH2 pKa = 7.07CSTPLLCSAVNNLPMGYYY20 pKa = 8.92VLQQPPIPATGQPHHH35 pKa = 7.19DDD37 pKa = 3.05MGISNCHHH45 pKa = 4.94VNGVPAPSNFQPMRR59 pKa = 11.84MNSGNDDD66 pKa = 2.79VMEEE70 pKa = 4.29NATNVTPAVPPTTTMSSVSEEE91 pKa = 4.19PVSPTSVASSGNFPFTASDDD111 pKa = 3.55SGMGVDDD118 pKa = 4.24SALDDD123 pKa = 3.47AFTTDDD129 pKa = 3.02ASSVGLQLGPDDD141 pKa = 3.28GAGNSRR147 pKa = 11.84DDD149 pKa = 3.58LRR151 pKa = 11.84SLDDD155 pKa = 3.67IQWNFSLTDDD165 pKa = 3.54TADDD169 pKa = 4.51SNLGDDD175 pKa = 4.41GALGNYYY182 pKa = 9.32GSPFLPSDDD191 pKa = 3.82EEE193 pKa = 4.11LLDDD197 pKa = 4.49PEEE200 pKa = 3.97EEE202 pKa = 4.86DD203 pKa = 3.6VEEE206 pKa = 4.18EE207 pKa = 4.25FVDDD211 pKa = 3.95VPGQPCSQSDDD222 pKa = 3.83EE223 pKa = 4.43EE224 pKa = 4.69KK225 pKa = 10.95
HHH2 pKa = 7.07CSTPLLCSAVNNLPMGYYY20 pKa = 8.92VLQQPPIPATGQPHHH35 pKa = 7.19DDD37 pKa = 3.05MGISNCHHH45 pKa = 4.94VNGVPAPSNFQPMRR59 pKa = 11.84MNSGNDDD66 pKa = 2.79VMEEE70 pKa = 4.29NATNVTPAVPPTTTMSSVSEEE91 pKa = 4.19PVSPTSVASSGNFPFTASDDD111 pKa = 3.55SGMGVDDD118 pKa = 4.24SALDDD123 pKa = 3.47AFTTDDD129 pKa = 3.02ASSVGLQLGPDDD141 pKa = 3.28GAGNSRR147 pKa = 11.84DDD149 pKa = 3.58LRR151 pKa = 11.84SLDDD155 pKa = 3.67IQWNFSLTDDD165 pKa = 3.54TADDD169 pKa = 4.51SNLGDDD175 pKa = 4.41GALGNYYY182 pKa = 9.32GSPFLPSDDD191 pKa = 3.82EEE193 pKa = 4.11LLDDD197 pKa = 4.49PEEE200 pKa = 3.97EEE202 pKa = 4.86DD203 pKa = 3.6VEEE206 pKa = 4.18EE207 pKa = 4.25FVDDD211 pKa = 3.95VPGQPCSQSDDD222 pKa = 3.83EE223 pKa = 4.43EE224 pKa = 4.69KK225 pKa = 10.95
Molecular weight: 23.44 kDa
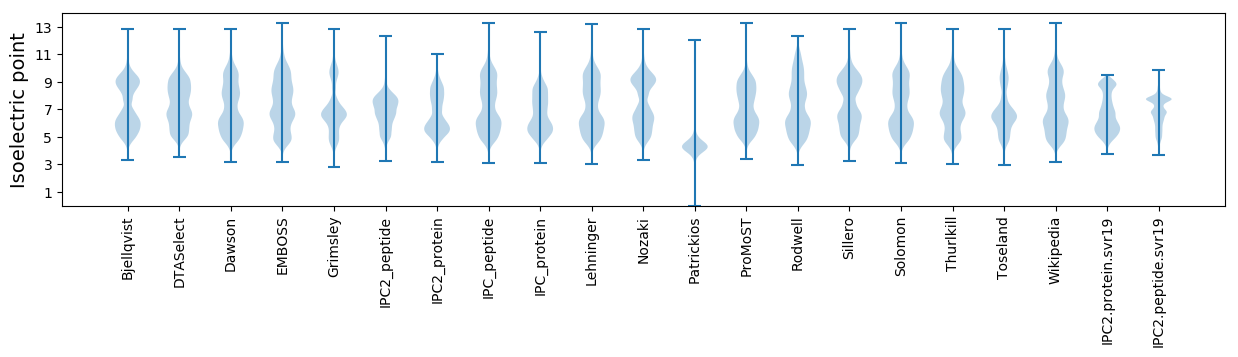
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6J0ZYK4|A0A6J0ZYK4_9ROSI uncharacterized protein LOC110413221 OS=Herrania umbratica OX=108875 GN=LOC110413221 PE=4 SV=1
MM1 pKa = 8.15RR2 pKa = 11.84DD3 pKa = 2.98PGARR7 pKa = 11.84RR8 pKa = 11.84GSSPAWAAGGEE19 pKa = 4.37LGRR22 pKa = 11.84APARR26 pKa = 11.84LAAARR31 pKa = 11.84AGAARR36 pKa = 11.84RR37 pKa = 11.84ARR39 pKa = 11.84PPRR42 pKa = 11.84PRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84APPRR50 pKa = 11.84GARR53 pKa = 11.84RR54 pKa = 11.84PEE56 pKa = 4.24GVVGAAPGAPGAARR70 pKa = 11.84RR71 pKa = 11.84AAPGPAAAGRR81 pKa = 11.84PGAAARR87 pKa = 11.84RR88 pKa = 11.84PPRR91 pKa = 11.84AAPARR96 pKa = 11.84SAGAGGAAAGGRR108 pKa = 11.84GRR110 pKa = 11.84RR111 pKa = 11.84AAAAPGRR118 pKa = 11.84GRR120 pKa = 11.84RR121 pKa = 11.84GGAAGAGGGPGRR133 pKa = 11.84ARR135 pKa = 11.84RR136 pKa = 11.84ARR138 pKa = 11.84AAAPGRR144 pKa = 11.84PPRR147 pKa = 11.84ACGAARR153 pKa = 11.84PGSRR157 pKa = 11.84LGAAAGPTSAGRR169 pKa = 11.84RR170 pKa = 11.84RR171 pKa = 11.84GAGGRR176 pKa = 11.84RR177 pKa = 11.84RR178 pKa = 11.84RR179 pKa = 11.84GSAPGRR185 pKa = 11.84RR186 pKa = 11.84AAGGGGGAGSGARR199 pKa = 11.84ARR201 pKa = 11.84RR202 pKa = 11.84PGRR205 pKa = 11.84GPAAPACARR214 pKa = 11.84RR215 pKa = 11.84CPAGAAGRR223 pKa = 11.84LLINLL228 pKa = 4.34
MM1 pKa = 8.15RR2 pKa = 11.84DD3 pKa = 2.98PGARR7 pKa = 11.84RR8 pKa = 11.84GSSPAWAAGGEE19 pKa = 4.37LGRR22 pKa = 11.84APARR26 pKa = 11.84LAAARR31 pKa = 11.84AGAARR36 pKa = 11.84RR37 pKa = 11.84ARR39 pKa = 11.84PPRR42 pKa = 11.84PRR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84APPRR50 pKa = 11.84GARR53 pKa = 11.84RR54 pKa = 11.84PEE56 pKa = 4.24GVVGAAPGAPGAARR70 pKa = 11.84RR71 pKa = 11.84AAPGPAAAGRR81 pKa = 11.84PGAAARR87 pKa = 11.84RR88 pKa = 11.84PPRR91 pKa = 11.84AAPARR96 pKa = 11.84SAGAGGAAAGGRR108 pKa = 11.84GRR110 pKa = 11.84RR111 pKa = 11.84AAAAPGRR118 pKa = 11.84GRR120 pKa = 11.84RR121 pKa = 11.84GGAAGAGGGPGRR133 pKa = 11.84ARR135 pKa = 11.84RR136 pKa = 11.84ARR138 pKa = 11.84AAAPGRR144 pKa = 11.84PPRR147 pKa = 11.84ACGAARR153 pKa = 11.84PGSRR157 pKa = 11.84LGAAAGPTSAGRR169 pKa = 11.84RR170 pKa = 11.84RR171 pKa = 11.84GAGGRR176 pKa = 11.84RR177 pKa = 11.84RR178 pKa = 11.84RR179 pKa = 11.84GSAPGRR185 pKa = 11.84RR186 pKa = 11.84AAGGGGGAGSGARR199 pKa = 11.84ARR201 pKa = 11.84RR202 pKa = 11.84PGRR205 pKa = 11.84GPAAPACARR214 pKa = 11.84RR215 pKa = 11.84CPAGAAGRR223 pKa = 11.84LLINLL228 pKa = 4.34
Molecular weight: 21.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
11398064 |
37 |
5457 |
477.9 |
53.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.819 ± 0.012 | 1.909 ± 0.007 |
5.237 ± 0.01 | 6.524 ± 0.016 |
4.229 ± 0.009 | 6.492 ± 0.012 |
2.349 ± 0.006 | 5.282 ± 0.009 |
6.106 ± 0.015 | 9.921 ± 0.021 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.402 ± 0.006 | 4.436 ± 0.01 |
4.857 ± 0.013 | 3.798 ± 0.012 |
5.264 ± 0.011 | 9.06 ± 0.017 |
4.752 ± 0.008 | 6.478 ± 0.01 |
1.275 ± 0.005 | 2.771 ± 0.009 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
