
Klebsiella pneumoniae subsp. pneumoniae DSM 30104 = JCM 1662 = NBRC 14940
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Klebsiella; Klebsiella pneumoniae; Klebsiella pneumoniae subsp. pneumoniae
Average proteome isoelectric point is 7.19
Get precalculated fractions of proteins
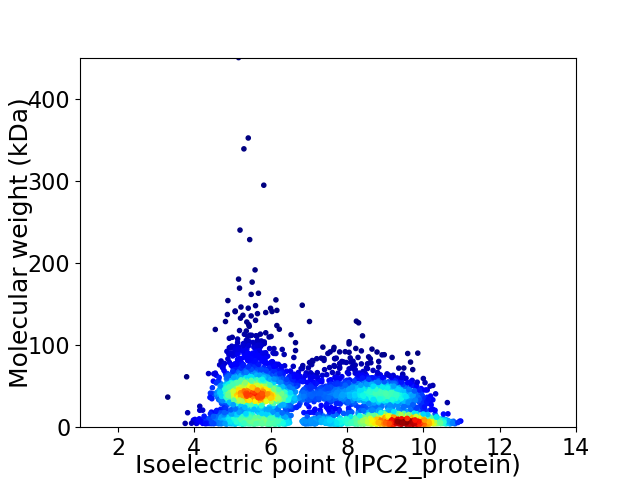
Virtual 2D-PAGE plot for 4528 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
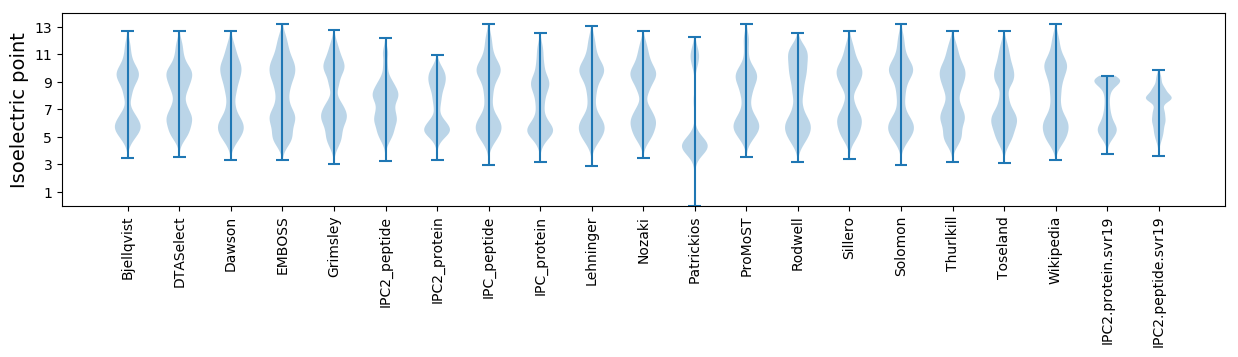
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J2DNB8|J2DNB8_KLEPN Transporter major facilitator family protein OS=Klebsiella pneumoniae subsp. pneumoniae DSM 30104 = JCM 1662 = NBRC 14940 OX=1162296 GN=UUU_12150 PE=4 SV=1
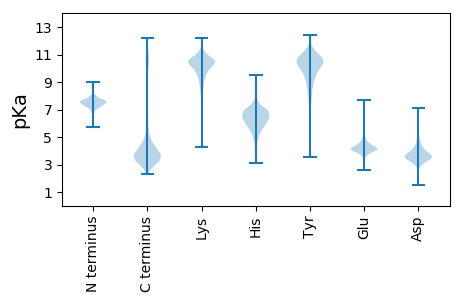
MM1 pKa = 7.46SGNIGANPVGVIKK14 pKa = 10.73DD15 pKa = 4.46GITATPTGFLGLSLEE30 pKa = 4.35YY31 pKa = 10.77AGGGEE36 pKa = 4.27FEE38 pKa = 5.29RR39 pKa = 11.84IGAGNYY45 pKa = 7.23MVVISSPTTEE55 pKa = 4.25GLIGSILNLNLGTSISMEE73 pKa = 4.05VTDD76 pKa = 3.36STKK79 pKa = 10.58YY80 pKa = 7.39VTGNVLHH87 pKa = 6.89TDD89 pKa = 2.98SDD91 pKa = 4.26GDD93 pKa = 3.74VGDD96 pKa = 4.09TGNEE100 pKa = 3.75ITVSSIQFGSQIEE113 pKa = 4.44TVTAGGVTINGTYY126 pKa = 8.21GTLIIKK132 pKa = 10.27SDD134 pKa = 3.27GSYY137 pKa = 10.47SYY139 pKa = 10.41HH140 pKa = 7.08ANGTTGVGQSDD151 pKa = 4.27VFTYY155 pKa = 10.04TITDD159 pKa = 3.88GLGHH163 pKa = 6.64TSSTTLTMNIEE174 pKa = 4.21STPPVAVDD182 pKa = 5.24DD183 pKa = 4.26IASLSAATAMQSVTWTQSVSNLAVPNTAVAANATSAAKK221 pKa = 8.88VTSQNFTIAEE231 pKa = 3.98GRR233 pKa = 11.84EE234 pKa = 3.73VDD236 pKa = 3.83GASITLNLSGNRR248 pKa = 11.84TSGLSLLNGDD258 pKa = 4.37LSGTVTLIHH267 pKa = 6.55HH268 pKa = 5.27VTVGGVTTDD277 pKa = 3.3VTVWTSSLTMALDD290 pKa = 4.26GSLLSPSSDD299 pKa = 2.91TDD301 pKa = 3.69TVTLNMVNGGIPLQLAAGDD320 pKa = 3.88YY321 pKa = 8.7TLSISHH327 pKa = 6.41TMHH330 pKa = 7.02AGTALSYY337 pKa = 11.03NLSASVAGTEE347 pKa = 4.1ILTDD351 pKa = 3.15AHH353 pKa = 5.99LTVGQSVAGNILDD366 pKa = 4.32GSDD369 pKa = 3.99ANGTPDD375 pKa = 3.43TLGSLHH381 pKa = 6.93SGFTISGEE389 pKa = 4.15NNLGIATNWTFHH401 pKa = 7.15SDD403 pKa = 3.17GTVTNDD409 pKa = 3.01TGTSASNGNAVLYY422 pKa = 10.62GEE424 pKa = 4.47YY425 pKa = 10.73GILTINGQGGYY436 pKa = 8.47TYY438 pKa = 10.82QLNGGVNTDD447 pKa = 4.27AITSKK452 pKa = 9.46EE453 pKa = 4.32TFTYY457 pKa = 10.44TLISSDD463 pKa = 3.81GGSSTANLTIDD474 pKa = 4.36LHH476 pKa = 5.23PQIAGSVNDD485 pKa = 4.66DD486 pKa = 3.97SVHH489 pKa = 4.89STAYY493 pKa = 10.62DD494 pKa = 3.36DD495 pKa = 3.91TFSMGVGADD504 pKa = 3.26TLVYY508 pKa = 10.96NLLADD513 pKa = 4.97DD514 pKa = 3.95NTGGNGSDD522 pKa = 2.74IWSDD526 pKa = 3.28FSVAQGDD533 pKa = 4.53HH534 pKa = 6.66IDD536 pKa = 3.59VSALLVGWNGSSDD549 pKa = 3.41TLGNYY554 pKa = 7.17ITLSYY559 pKa = 10.98VGGNTVVSIDD569 pKa = 3.69RR570 pKa = 11.84DD571 pKa = 3.8GTGGNTHH578 pKa = 6.04QPATLITLQGVHH590 pKa = 6.26INSLDD595 pKa = 3.72EE596 pKa = 6.3LIDD599 pKa = 3.88TNNSNN604 pKa = 3.77
MM1 pKa = 7.46SGNIGANPVGVIKK14 pKa = 10.73DD15 pKa = 4.46GITATPTGFLGLSLEE30 pKa = 4.35YY31 pKa = 10.77AGGGEE36 pKa = 4.27FEE38 pKa = 5.29RR39 pKa = 11.84IGAGNYY45 pKa = 7.23MVVISSPTTEE55 pKa = 4.25GLIGSILNLNLGTSISMEE73 pKa = 4.05VTDD76 pKa = 3.36STKK79 pKa = 10.58YY80 pKa = 7.39VTGNVLHH87 pKa = 6.89TDD89 pKa = 2.98SDD91 pKa = 4.26GDD93 pKa = 3.74VGDD96 pKa = 4.09TGNEE100 pKa = 3.75ITVSSIQFGSQIEE113 pKa = 4.44TVTAGGVTINGTYY126 pKa = 8.21GTLIIKK132 pKa = 10.27SDD134 pKa = 3.27GSYY137 pKa = 10.47SYY139 pKa = 10.41HH140 pKa = 7.08ANGTTGVGQSDD151 pKa = 4.27VFTYY155 pKa = 10.04TITDD159 pKa = 3.88GLGHH163 pKa = 6.64TSSTTLTMNIEE174 pKa = 4.21STPPVAVDD182 pKa = 5.24DD183 pKa = 4.26IASLSAATAMQSVTWTQSVSNLAVPNTAVAANATSAAKK221 pKa = 8.88VTSQNFTIAEE231 pKa = 3.98GRR233 pKa = 11.84EE234 pKa = 3.73VDD236 pKa = 3.83GASITLNLSGNRR248 pKa = 11.84TSGLSLLNGDD258 pKa = 4.37LSGTVTLIHH267 pKa = 6.55HH268 pKa = 5.27VTVGGVTTDD277 pKa = 3.3VTVWTSSLTMALDD290 pKa = 4.26GSLLSPSSDD299 pKa = 2.91TDD301 pKa = 3.69TVTLNMVNGGIPLQLAAGDD320 pKa = 3.88YY321 pKa = 8.7TLSISHH327 pKa = 6.41TMHH330 pKa = 7.02AGTALSYY337 pKa = 11.03NLSASVAGTEE347 pKa = 4.1ILTDD351 pKa = 3.15AHH353 pKa = 5.99LTVGQSVAGNILDD366 pKa = 4.32GSDD369 pKa = 3.99ANGTPDD375 pKa = 3.43TLGSLHH381 pKa = 6.93SGFTISGEE389 pKa = 4.15NNLGIATNWTFHH401 pKa = 7.15SDD403 pKa = 3.17GTVTNDD409 pKa = 3.01TGTSASNGNAVLYY422 pKa = 10.62GEE424 pKa = 4.47YY425 pKa = 10.73GILTINGQGGYY436 pKa = 8.47TYY438 pKa = 10.82QLNGGVNTDD447 pKa = 4.27AITSKK452 pKa = 9.46EE453 pKa = 4.32TFTYY457 pKa = 10.44TLISSDD463 pKa = 3.81GGSSTANLTIDD474 pKa = 4.36LHH476 pKa = 5.23PQIAGSVNDD485 pKa = 4.66DD486 pKa = 3.97SVHH489 pKa = 4.89STAYY493 pKa = 10.62DD494 pKa = 3.36DD495 pKa = 3.91TFSMGVGADD504 pKa = 3.26TLVYY508 pKa = 10.96NLLADD513 pKa = 4.97DD514 pKa = 3.95NTGGNGSDD522 pKa = 2.74IWSDD526 pKa = 3.28FSVAQGDD533 pKa = 4.53HH534 pKa = 6.66IDD536 pKa = 3.59VSALLVGWNGSSDD549 pKa = 3.41TLGNYY554 pKa = 7.17ITLSYY559 pKa = 10.98VGGNTVVSIDD569 pKa = 3.69RR570 pKa = 11.84DD571 pKa = 3.8GTGGNTHH578 pKa = 6.04QPATLITLQGVHH590 pKa = 6.26INSLDD595 pKa = 3.72EE596 pKa = 6.3LIDD599 pKa = 3.88TNNSNN604 pKa = 3.77
Molecular weight: 61.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|J2LLH4|J2LLH4_KLEPN Uncharacterized protein OS=Klebsiella pneumoniae subsp. pneumoniae DSM 30104 = JCM 1662 = NBRC 14940 OX=1162296 GN=UUU_36740 PE=4 SV=1
MM1 pKa = 7.6IRR3 pKa = 11.84LTLKK7 pKa = 10.43RR8 pKa = 11.84ATLVTSWSRR17 pKa = 11.84RR18 pKa = 11.84VKK20 pKa = 10.83LFLLPRR26 pKa = 11.84KK27 pKa = 7.36TLNSSKK33 pKa = 10.62HH34 pKa = 5.49VVLNWKK40 pKa = 9.38PNWLTFWQQLKK51 pKa = 10.49LVLSRR56 pKa = 11.84STPWNPP62 pKa = 3.15
MM1 pKa = 7.6IRR3 pKa = 11.84LTLKK7 pKa = 10.43RR8 pKa = 11.84ATLVTSWSRR17 pKa = 11.84RR18 pKa = 11.84VKK20 pKa = 10.83LFLLPRR26 pKa = 11.84KK27 pKa = 7.36TLNSSKK33 pKa = 10.62HH34 pKa = 5.49VVLNWKK40 pKa = 9.38PNWLTFWQQLKK51 pKa = 10.49LVLSRR56 pKa = 11.84STPWNPP62 pKa = 3.15
Molecular weight: 7.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1291935 |
37 |
4234 |
285.3 |
31.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.138 ± 0.049 | 1.089 ± 0.014 |
5.036 ± 0.035 | 5.172 ± 0.038 |
4.007 ± 0.03 | 7.459 ± 0.04 |
2.527 ± 0.026 | 5.717 ± 0.038 |
3.972 ± 0.04 | 10.86 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.707 ± 0.02 | 3.642 ± 0.028 |
4.659 ± 0.03 | 4.744 ± 0.034 |
6.159 ± 0.041 | 5.798 ± 0.035 |
5.23 ± 0.032 | 6.905 ± 0.033 |
1.474 ± 0.02 | 2.705 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
