
Methylobrevis pamukkalensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Pleomorphomonadaceae; Methylobrevis
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
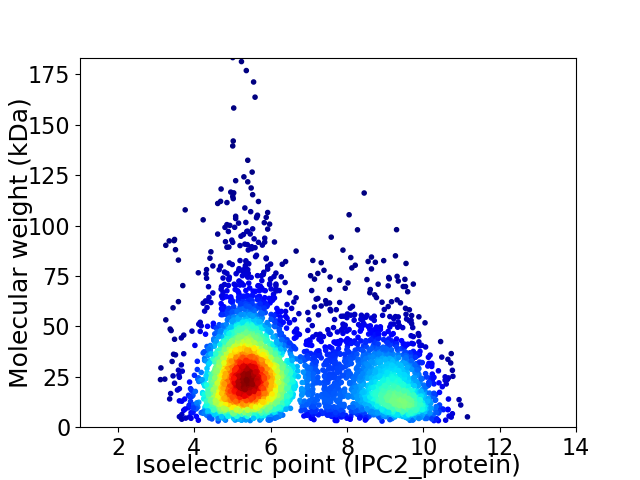
Virtual 2D-PAGE plot for 4503 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1E3H8J4|A0A1E3H8J4_9RHIZ Leucine--tRNA ligase OS=Methylobrevis pamukkalensis OX=1439726 GN=leuS PE=3 SV=1
MM1 pKa = 7.77DD2 pKa = 5.28HH3 pKa = 7.16LFGGNEE9 pKa = 3.92GDD11 pKa = 3.91FLSGGSDD18 pKa = 3.76DD19 pKa = 3.7DD20 pKa = 3.97TLYY23 pKa = 11.57GEE25 pKa = 5.13AGNDD29 pKa = 3.61TLDD32 pKa = 3.75GGSGYY37 pKa = 8.73DD38 pKa = 3.69TLHH41 pKa = 6.65GGAGKK46 pKa = 10.38DD47 pKa = 3.44KK48 pKa = 10.77LYY50 pKa = 11.29GGADD54 pKa = 3.21SDD56 pKa = 3.86TYY58 pKa = 11.04YY59 pKa = 10.63INAAEE64 pKa = 4.05GDD66 pKa = 4.02EE67 pKa = 4.41VYY69 pKa = 11.12EE70 pKa = 4.12NANGGLDD77 pKa = 3.4HH78 pKa = 6.95VFVTGSLYY86 pKa = 10.35TLTANVEE93 pKa = 3.93RR94 pKa = 11.84GTLYY98 pKa = 10.3TAGGTLTGNAEE109 pKa = 4.08NNHH112 pKa = 6.05LFGSTGNEE120 pKa = 3.58TLYY123 pKa = 11.43GLGGHH128 pKa = 6.55DD129 pKa = 4.18TLDD132 pKa = 3.61GGIGADD138 pKa = 3.46TFYY141 pKa = 11.36GGAGDD146 pKa = 3.75DD147 pKa = 3.71TFIVEE152 pKa = 4.22NVGDD156 pKa = 4.11KK157 pKa = 10.74VIEE160 pKa = 4.03YY161 pKa = 10.5SGGGVDD167 pKa = 4.45TVITQLGNYY176 pKa = 7.4TLGDD180 pKa = 3.44HH181 pKa = 6.77VEE183 pKa = 3.96NLKK186 pKa = 10.7RR187 pKa = 11.84GNDD190 pKa = 3.82GAGKK194 pKa = 8.8LTGNGGANTITGNINGDD211 pKa = 4.08TIDD214 pKa = 4.22GGAGQDD220 pKa = 3.48VMIGGKK226 pKa = 10.64GNDD229 pKa = 3.66FYY231 pKa = 11.86YY232 pKa = 10.98VDD234 pKa = 4.79DD235 pKa = 5.52LGDD238 pKa = 3.91SVIEE242 pKa = 4.12KK243 pKa = 10.37AGEE246 pKa = 4.0GLDD249 pKa = 3.45EE250 pKa = 4.5VFASTAVYY258 pKa = 10.43TMTDD262 pKa = 2.94HH263 pKa = 7.55VEE265 pKa = 4.14QLHH268 pKa = 4.41VTYY271 pKa = 10.17TGGQISYY278 pKa = 11.43GNGIDD283 pKa = 3.4NWIDD287 pKa = 3.42GNTGNDD293 pKa = 3.14QMFGYY298 pKa = 10.26AGKK301 pKa = 10.28DD302 pKa = 3.18KK303 pKa = 11.12LVGEE307 pKa = 4.9GGDD310 pKa = 3.56DD311 pKa = 3.66LLYY314 pKa = 11.09GGSEE318 pKa = 4.33DD319 pKa = 4.43DD320 pKa = 4.81ALDD323 pKa = 3.9GGEE326 pKa = 5.56GEE328 pKa = 5.03DD329 pKa = 4.86LLDD332 pKa = 4.51GGTGADD338 pKa = 3.6TMLGGIGDD346 pKa = 4.26DD347 pKa = 3.45TYY349 pKa = 11.22MVDD352 pKa = 3.6DD353 pKa = 5.3AGDD356 pKa = 3.73KK357 pKa = 9.86VVEE360 pKa = 4.25MPDD363 pKa = 3.08GGFDD367 pKa = 3.5VVNASTSYY375 pKa = 10.38TLGNYY380 pKa = 9.36QIEE383 pKa = 4.33EE384 pKa = 4.2LVLTGANAIDD394 pKa = 3.79GTGNGGINHH403 pKa = 6.43IVGNNAANVLDD414 pKa = 4.22GQFGADD420 pKa = 3.27TLEE423 pKa = 4.54GKK425 pKa = 9.89DD426 pKa = 3.71GNDD429 pKa = 3.09TYY431 pKa = 11.83VVDD434 pKa = 3.88NVGDD438 pKa = 3.97KK439 pKa = 10.23TVEE442 pKa = 4.04TADD445 pKa = 3.41QGIDD449 pKa = 3.31TVLASASFSLKK460 pKa = 9.76GQYY463 pKa = 9.47VEE465 pKa = 5.18NLTLTGDD472 pKa = 3.55GDD474 pKa = 4.44INGTGNKK481 pKa = 9.58LDD483 pKa = 3.69NTIKK487 pKa = 10.99GNGAANVLDD496 pKa = 4.2GAEE499 pKa = 4.39GADD502 pKa = 3.74SLEE505 pKa = 4.52GGAGADD511 pKa = 2.87QFVFSTKK518 pKa = 10.34LGASNIDD525 pKa = 3.59TIVDD529 pKa = 4.34FKK531 pKa = 11.6AVDD534 pKa = 3.65DD535 pKa = 4.91TILLDD540 pKa = 3.82DD541 pKa = 6.01AIFKK545 pKa = 9.63TLDD548 pKa = 3.38LGALAAGEE556 pKa = 4.04FRR558 pKa = 11.84IGAAAKK564 pKa = 10.39DD565 pKa = 3.75ADD567 pKa = 4.01DD568 pKa = 3.77HH569 pKa = 7.0VIYY572 pKa = 10.68AKK574 pKa = 9.81GTGALYY580 pKa = 10.96YY581 pKa = 10.75DD582 pKa = 4.38ADD584 pKa = 4.16GNGAGAAIQFATLDD598 pKa = 3.6TGLSLTSADD607 pKa = 3.71FLVII611 pKa = 4.12
MM1 pKa = 7.77DD2 pKa = 5.28HH3 pKa = 7.16LFGGNEE9 pKa = 3.92GDD11 pKa = 3.91FLSGGSDD18 pKa = 3.76DD19 pKa = 3.7DD20 pKa = 3.97TLYY23 pKa = 11.57GEE25 pKa = 5.13AGNDD29 pKa = 3.61TLDD32 pKa = 3.75GGSGYY37 pKa = 8.73DD38 pKa = 3.69TLHH41 pKa = 6.65GGAGKK46 pKa = 10.38DD47 pKa = 3.44KK48 pKa = 10.77LYY50 pKa = 11.29GGADD54 pKa = 3.21SDD56 pKa = 3.86TYY58 pKa = 11.04YY59 pKa = 10.63INAAEE64 pKa = 4.05GDD66 pKa = 4.02EE67 pKa = 4.41VYY69 pKa = 11.12EE70 pKa = 4.12NANGGLDD77 pKa = 3.4HH78 pKa = 6.95VFVTGSLYY86 pKa = 10.35TLTANVEE93 pKa = 3.93RR94 pKa = 11.84GTLYY98 pKa = 10.3TAGGTLTGNAEE109 pKa = 4.08NNHH112 pKa = 6.05LFGSTGNEE120 pKa = 3.58TLYY123 pKa = 11.43GLGGHH128 pKa = 6.55DD129 pKa = 4.18TLDD132 pKa = 3.61GGIGADD138 pKa = 3.46TFYY141 pKa = 11.36GGAGDD146 pKa = 3.75DD147 pKa = 3.71TFIVEE152 pKa = 4.22NVGDD156 pKa = 4.11KK157 pKa = 10.74VIEE160 pKa = 4.03YY161 pKa = 10.5SGGGVDD167 pKa = 4.45TVITQLGNYY176 pKa = 7.4TLGDD180 pKa = 3.44HH181 pKa = 6.77VEE183 pKa = 3.96NLKK186 pKa = 10.7RR187 pKa = 11.84GNDD190 pKa = 3.82GAGKK194 pKa = 8.8LTGNGGANTITGNINGDD211 pKa = 4.08TIDD214 pKa = 4.22GGAGQDD220 pKa = 3.48VMIGGKK226 pKa = 10.64GNDD229 pKa = 3.66FYY231 pKa = 11.86YY232 pKa = 10.98VDD234 pKa = 4.79DD235 pKa = 5.52LGDD238 pKa = 3.91SVIEE242 pKa = 4.12KK243 pKa = 10.37AGEE246 pKa = 4.0GLDD249 pKa = 3.45EE250 pKa = 4.5VFASTAVYY258 pKa = 10.43TMTDD262 pKa = 2.94HH263 pKa = 7.55VEE265 pKa = 4.14QLHH268 pKa = 4.41VTYY271 pKa = 10.17TGGQISYY278 pKa = 11.43GNGIDD283 pKa = 3.4NWIDD287 pKa = 3.42GNTGNDD293 pKa = 3.14QMFGYY298 pKa = 10.26AGKK301 pKa = 10.28DD302 pKa = 3.18KK303 pKa = 11.12LVGEE307 pKa = 4.9GGDD310 pKa = 3.56DD311 pKa = 3.66LLYY314 pKa = 11.09GGSEE318 pKa = 4.33DD319 pKa = 4.43DD320 pKa = 4.81ALDD323 pKa = 3.9GGEE326 pKa = 5.56GEE328 pKa = 5.03DD329 pKa = 4.86LLDD332 pKa = 4.51GGTGADD338 pKa = 3.6TMLGGIGDD346 pKa = 4.26DD347 pKa = 3.45TYY349 pKa = 11.22MVDD352 pKa = 3.6DD353 pKa = 5.3AGDD356 pKa = 3.73KK357 pKa = 9.86VVEE360 pKa = 4.25MPDD363 pKa = 3.08GGFDD367 pKa = 3.5VVNASTSYY375 pKa = 10.38TLGNYY380 pKa = 9.36QIEE383 pKa = 4.33EE384 pKa = 4.2LVLTGANAIDD394 pKa = 3.79GTGNGGINHH403 pKa = 6.43IVGNNAANVLDD414 pKa = 4.22GQFGADD420 pKa = 3.27TLEE423 pKa = 4.54GKK425 pKa = 9.89DD426 pKa = 3.71GNDD429 pKa = 3.09TYY431 pKa = 11.83VVDD434 pKa = 3.88NVGDD438 pKa = 3.97KK439 pKa = 10.23TVEE442 pKa = 4.04TADD445 pKa = 3.41QGIDD449 pKa = 3.31TVLASASFSLKK460 pKa = 9.76GQYY463 pKa = 9.47VEE465 pKa = 5.18NLTLTGDD472 pKa = 3.55GDD474 pKa = 4.44INGTGNKK481 pKa = 9.58LDD483 pKa = 3.69NTIKK487 pKa = 10.99GNGAANVLDD496 pKa = 4.2GAEE499 pKa = 4.39GADD502 pKa = 3.74SLEE505 pKa = 4.52GGAGADD511 pKa = 2.87QFVFSTKK518 pKa = 10.34LGASNIDD525 pKa = 3.59TIVDD529 pKa = 4.34FKK531 pKa = 11.6AVDD534 pKa = 3.65DD535 pKa = 4.91TILLDD540 pKa = 3.82DD541 pKa = 6.01AIFKK545 pKa = 9.63TLDD548 pKa = 3.38LGALAAGEE556 pKa = 4.04FRR558 pKa = 11.84IGAAAKK564 pKa = 10.39DD565 pKa = 3.75ADD567 pKa = 4.01DD568 pKa = 3.77HH569 pKa = 7.0VIYY572 pKa = 10.68AKK574 pKa = 9.81GTGALYY580 pKa = 10.96YY581 pKa = 10.75DD582 pKa = 4.38ADD584 pKa = 4.16GNGAGAAIQFATLDD598 pKa = 3.6TGLSLTSADD607 pKa = 3.71FLVII611 pKa = 4.12
Molecular weight: 62.25 kDa
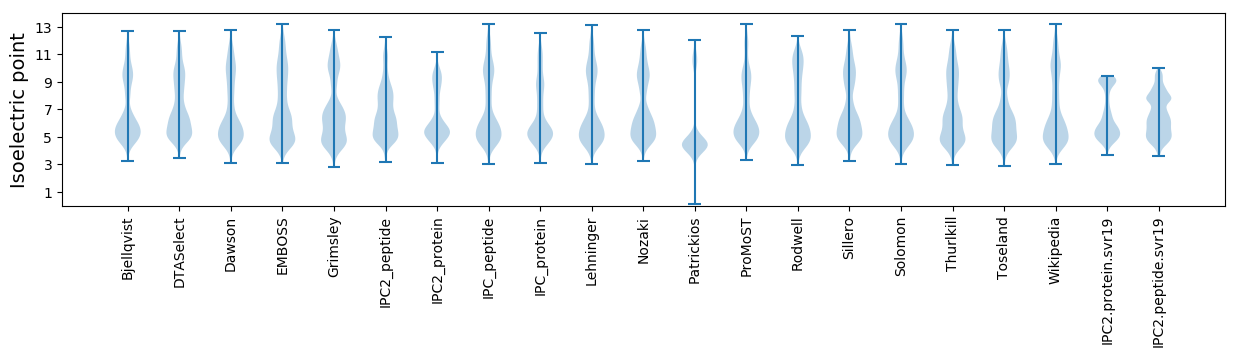
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1E3H6A7|A0A1E3H6A7_9RHIZ Histidine kinase OS=Methylobrevis pamukkalensis OX=1439726 GN=envZ_2 PE=4 SV=1
MM1 pKa = 7.11TATVTAPLSIAVSSTPSVFRR21 pKa = 11.84QRR23 pKa = 11.84PVRR26 pKa = 11.84QVQSQSSTHH35 pKa = 6.06LFAVGQAVRR44 pKa = 11.84LKK46 pKa = 10.86GGFGHH51 pKa = 7.53PSQFAGASTSPPRR64 pKa = 11.84CRR66 pKa = 11.84RR67 pKa = 11.84AATRR71 pKa = 11.84PSIASATTTNATSASRR87 pKa = 11.84RR88 pKa = 11.84RR89 pKa = 11.84KK90 pKa = 9.76ASRR93 pKa = 11.84LPAPPLTVTARR104 pKa = 11.84II105 pKa = 3.84
MM1 pKa = 7.11TATVTAPLSIAVSSTPSVFRR21 pKa = 11.84QRR23 pKa = 11.84PVRR26 pKa = 11.84QVQSQSSTHH35 pKa = 6.06LFAVGQAVRR44 pKa = 11.84LKK46 pKa = 10.86GGFGHH51 pKa = 7.53PSQFAGASTSPPRR64 pKa = 11.84CRR66 pKa = 11.84RR67 pKa = 11.84AATRR71 pKa = 11.84PSIASATTTNATSASRR87 pKa = 11.84RR88 pKa = 11.84RR89 pKa = 11.84KK90 pKa = 9.76ASRR93 pKa = 11.84LPAPPLTVTARR104 pKa = 11.84II105 pKa = 3.84
Molecular weight: 11.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1213789 |
29 |
1730 |
269.6 |
28.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.644 ± 0.059 | 0.871 ± 0.013 |
6.099 ± 0.037 | 5.486 ± 0.03 |
3.53 ± 0.023 | 9.108 ± 0.044 |
1.985 ± 0.022 | 4.805 ± 0.033 |
2.519 ± 0.029 | 9.904 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.422 ± 0.019 | 2.061 ± 0.019 |
5.574 ± 0.033 | 2.409 ± 0.02 |
7.989 ± 0.053 | 5.184 ± 0.027 |
5.466 ± 0.03 | 7.787 ± 0.032 |
1.259 ± 0.016 | 1.899 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
