
Phocoena phocoena papillomavirus 2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Upsilonpapillomavirus; Upsilonpapillomavirus 3
Average proteome isoelectric point is 5.8
Get precalculated fractions of proteins
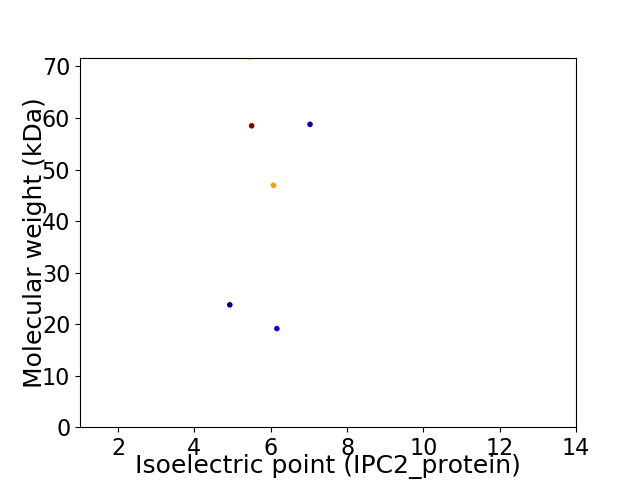
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
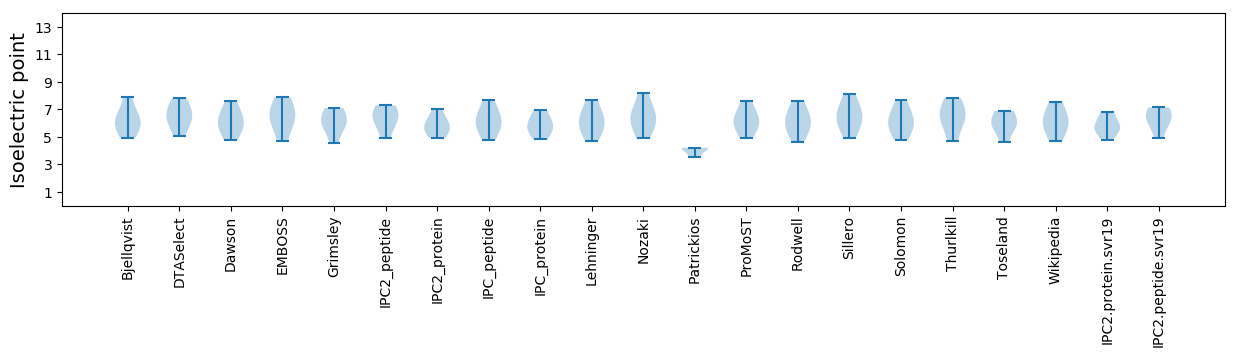
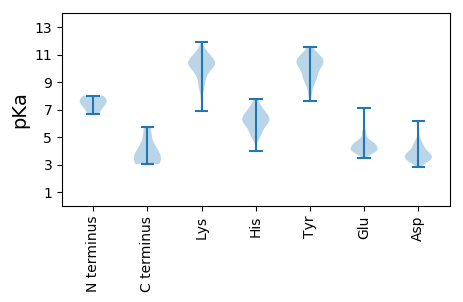
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F2VIR3|F2VIR3_9PAPI Replication protein E1 OS=Phocoena phocoena papillomavirus 2 OX=706526 GN=E1 PE=3 SV=1
MM1 pKa = 7.78ADD3 pKa = 3.67DD4 pKa = 4.43PRR6 pKa = 11.84TIKK9 pKa = 10.46DD10 pKa = 3.16LCKK13 pKa = 10.86NFDD16 pKa = 3.97LSVQALLILCVFCKK30 pKa = 10.45KK31 pKa = 10.25QLTDD35 pKa = 3.52LEE37 pKa = 4.39IWSFSHH43 pKa = 5.92KK44 pKa = 7.93QLRR47 pKa = 11.84VVWKK51 pKa = 10.33KK52 pKa = 10.13GFPFAACKK60 pKa = 10.25KK61 pKa = 8.81CLEE64 pKa = 4.07VCALVDD70 pKa = 3.24CWRR73 pKa = 11.84NFQRR77 pKa = 11.84SATAKK82 pKa = 10.0QVEE85 pKa = 4.6EE86 pKa = 4.32DD87 pKa = 3.27TGKK90 pKa = 10.85ALGDD94 pKa = 3.51LDD96 pKa = 4.35VRR98 pKa = 11.84CMGCYY103 pKa = 10.7KK104 pKa = 11.03LMTTLEE110 pKa = 4.07KK111 pKa = 10.35AYY113 pKa = 10.31QLEE116 pKa = 4.59DD117 pKa = 3.12EE118 pKa = 4.67KK119 pKa = 11.16QFCKK123 pKa = 10.23IAGYY127 pKa = 9.07WKK129 pKa = 10.21GYY131 pKa = 9.78CINCLTDD138 pKa = 3.71PPPLVSWFNYY148 pKa = 8.78AVTVGGPPPLITWGFDD164 pKa = 3.23PPRR167 pKa = 11.84RR168 pKa = 11.84PAQSVSSGSSWTITTSTSSSGRR190 pKa = 11.84EE191 pKa = 3.98PEE193 pKa = 4.51PDD195 pKa = 3.68DD196 pKa = 5.56PEE198 pKa = 7.13DD199 pKa = 4.68GDD201 pKa = 4.56SSDD204 pKa = 4.42DD205 pKa = 3.73QEE207 pKa = 4.41EE208 pKa = 4.06MLII211 pKa = 4.27
MM1 pKa = 7.78ADD3 pKa = 3.67DD4 pKa = 4.43PRR6 pKa = 11.84TIKK9 pKa = 10.46DD10 pKa = 3.16LCKK13 pKa = 10.86NFDD16 pKa = 3.97LSVQALLILCVFCKK30 pKa = 10.45KK31 pKa = 10.25QLTDD35 pKa = 3.52LEE37 pKa = 4.39IWSFSHH43 pKa = 5.92KK44 pKa = 7.93QLRR47 pKa = 11.84VVWKK51 pKa = 10.33KK52 pKa = 10.13GFPFAACKK60 pKa = 10.25KK61 pKa = 8.81CLEE64 pKa = 4.07VCALVDD70 pKa = 3.24CWRR73 pKa = 11.84NFQRR77 pKa = 11.84SATAKK82 pKa = 10.0QVEE85 pKa = 4.6EE86 pKa = 4.32DD87 pKa = 3.27TGKK90 pKa = 10.85ALGDD94 pKa = 3.51LDD96 pKa = 4.35VRR98 pKa = 11.84CMGCYY103 pKa = 10.7KK104 pKa = 11.03LMTTLEE110 pKa = 4.07KK111 pKa = 10.35AYY113 pKa = 10.31QLEE116 pKa = 4.59DD117 pKa = 3.12EE118 pKa = 4.67KK119 pKa = 11.16QFCKK123 pKa = 10.23IAGYY127 pKa = 9.07WKK129 pKa = 10.21GYY131 pKa = 9.78CINCLTDD138 pKa = 3.71PPPLVSWFNYY148 pKa = 8.78AVTVGGPPPLITWGFDD164 pKa = 3.23PPRR167 pKa = 11.84RR168 pKa = 11.84PAQSVSSGSSWTITTSTSSSGRR190 pKa = 11.84EE191 pKa = 3.98PEE193 pKa = 4.51PDD195 pKa = 3.68DD196 pKa = 5.56PEE198 pKa = 7.13DD199 pKa = 4.68GDD201 pKa = 4.56SSDD204 pKa = 4.42DD205 pKa = 3.73QEE207 pKa = 4.41EE208 pKa = 4.06MLII211 pKa = 4.27
Molecular weight: 23.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F2VIR7|F2VIR7_9PAPI Major capsid protein L1 OS=Phocoena phocoena papillomavirus 2 OX=706526 GN=L1 PE=3 SV=1
MM1 pKa = 7.62PLILVVAFTYY11 pKa = 10.26IHH13 pKa = 6.79IFAGRR18 pKa = 11.84NANACIFNIFQMLHH32 pKa = 6.55LPPPAPLSRR41 pKa = 11.84VLHH44 pKa = 5.73TDD46 pKa = 3.04EE47 pKa = 4.63FVSRR51 pKa = 11.84TSTFYY56 pKa = 10.02HH57 pKa = 6.15CQSEE61 pKa = 4.12RR62 pKa = 11.84LITIGHH68 pKa = 7.38PFYY71 pKa = 10.82KK72 pKa = 10.53VQDD75 pKa = 3.9GGNVVAEE82 pKa = 4.13KK83 pKa = 10.73VSPNQYY89 pKa = 9.42RR90 pKa = 11.84VFRR93 pKa = 11.84VTLPDD98 pKa = 4.27PNVLALTDD106 pKa = 3.48SSLYY110 pKa = 10.46NPEE113 pKa = 3.88KK114 pKa = 10.63EE115 pKa = 3.85RR116 pKa = 11.84LVWILRR122 pKa = 11.84GIDD125 pKa = 3.39VGRR128 pKa = 11.84GGPLGIGVTGHH139 pKa = 7.4PYY141 pKa = 10.66LDD143 pKa = 3.52KK144 pKa = 11.41LKK146 pKa = 10.88DD147 pKa = 3.51AEE149 pKa = 4.51NPNGSYY155 pKa = 11.2NKK157 pKa = 10.32GSDD160 pKa = 3.42DD161 pKa = 3.7ARR163 pKa = 11.84QNVCMDD169 pKa = 4.32PKK171 pKa = 10.31SVQMLILGCAPAIGQHH187 pKa = 4.91WDD189 pKa = 3.15KK190 pKa = 11.92AEE192 pKa = 4.06ACASDD197 pKa = 4.05RR198 pKa = 11.84DD199 pKa = 4.17AVDD202 pKa = 3.32ACPPLEE208 pKa = 4.59LKK210 pKa = 9.19NTTIEE215 pKa = 4.51DD216 pKa = 3.36GDD218 pKa = 3.81MFDD221 pKa = 5.32IGFGCMNNSTLQATFSAVPLDD242 pKa = 3.49ITNTTTKK249 pKa = 10.54HH250 pKa = 5.62PDD252 pKa = 3.03ILKK255 pKa = 7.88MTGDD259 pKa = 3.85IYY261 pKa = 11.54GNTCWFCVSRR271 pKa = 11.84EE272 pKa = 3.96QMFARR277 pKa = 11.84HH278 pKa = 5.04LWSRR282 pKa = 11.84NGDD285 pKa = 3.55NGDD288 pKa = 4.16PVPHH292 pKa = 6.87TDD294 pKa = 2.97GHH296 pKa = 6.76KK297 pKa = 10.09DD298 pKa = 3.0TSLYY302 pKa = 10.94LSGTGDD308 pKa = 3.97TKK310 pKa = 11.15TMATPVYY317 pKa = 9.7FNTPSGSLLTSEE329 pKa = 4.25TQLFNRR335 pKa = 11.84PFWMQRR341 pKa = 11.84AQGLNNGVCWNNTLFVTVADD361 pKa = 3.92NTRR364 pKa = 11.84GTNLNISVAKK374 pKa = 10.43QEE376 pKa = 4.21GKK378 pKa = 9.97NAEE381 pKa = 4.41SYY383 pKa = 8.1TASNYY388 pKa = 8.41KK389 pKa = 10.07HH390 pKa = 6.24YY391 pKa = 10.73NRR393 pKa = 11.84HH394 pKa = 5.18CEE396 pKa = 3.68IYY398 pKa = 9.47EE399 pKa = 4.11LEE401 pKa = 5.3FILQLATVALTPEE414 pKa = 4.56ALSHH418 pKa = 5.61LHH420 pKa = 7.03AMDD423 pKa = 3.72STILTKK429 pKa = 10.34WNLGFTASNATAVEE443 pKa = 3.98STYY446 pKa = 10.75RR447 pKa = 11.84YY448 pKa = 9.73INSKK452 pKa = 8.25ATKK455 pKa = 9.69CPIDD459 pKa = 3.64IPEE462 pKa = 4.84PEE464 pKa = 4.99PKK466 pKa = 10.1DD467 pKa = 3.44KK468 pKa = 11.46YY469 pKa = 11.25ADD471 pKa = 3.49LTFWNVDD478 pKa = 3.21VSKK481 pKa = 11.0SLSKK485 pKa = 10.78DD486 pKa = 3.23LSAFPLGRR494 pKa = 11.84KK495 pKa = 8.68FLYY498 pKa = 10.02QAGLQNGATRR508 pKa = 11.84SATKK512 pKa = 10.4RR513 pKa = 11.84SATKK517 pKa = 10.06TSTKK521 pKa = 10.31VPTKK525 pKa = 10.1RR526 pKa = 11.84KK527 pKa = 9.53RR528 pKa = 11.84RR529 pKa = 3.71
MM1 pKa = 7.62PLILVVAFTYY11 pKa = 10.26IHH13 pKa = 6.79IFAGRR18 pKa = 11.84NANACIFNIFQMLHH32 pKa = 6.55LPPPAPLSRR41 pKa = 11.84VLHH44 pKa = 5.73TDD46 pKa = 3.04EE47 pKa = 4.63FVSRR51 pKa = 11.84TSTFYY56 pKa = 10.02HH57 pKa = 6.15CQSEE61 pKa = 4.12RR62 pKa = 11.84LITIGHH68 pKa = 7.38PFYY71 pKa = 10.82KK72 pKa = 10.53VQDD75 pKa = 3.9GGNVVAEE82 pKa = 4.13KK83 pKa = 10.73VSPNQYY89 pKa = 9.42RR90 pKa = 11.84VFRR93 pKa = 11.84VTLPDD98 pKa = 4.27PNVLALTDD106 pKa = 3.48SSLYY110 pKa = 10.46NPEE113 pKa = 3.88KK114 pKa = 10.63EE115 pKa = 3.85RR116 pKa = 11.84LVWILRR122 pKa = 11.84GIDD125 pKa = 3.39VGRR128 pKa = 11.84GGPLGIGVTGHH139 pKa = 7.4PYY141 pKa = 10.66LDD143 pKa = 3.52KK144 pKa = 11.41LKK146 pKa = 10.88DD147 pKa = 3.51AEE149 pKa = 4.51NPNGSYY155 pKa = 11.2NKK157 pKa = 10.32GSDD160 pKa = 3.42DD161 pKa = 3.7ARR163 pKa = 11.84QNVCMDD169 pKa = 4.32PKK171 pKa = 10.31SVQMLILGCAPAIGQHH187 pKa = 4.91WDD189 pKa = 3.15KK190 pKa = 11.92AEE192 pKa = 4.06ACASDD197 pKa = 4.05RR198 pKa = 11.84DD199 pKa = 4.17AVDD202 pKa = 3.32ACPPLEE208 pKa = 4.59LKK210 pKa = 9.19NTTIEE215 pKa = 4.51DD216 pKa = 3.36GDD218 pKa = 3.81MFDD221 pKa = 5.32IGFGCMNNSTLQATFSAVPLDD242 pKa = 3.49ITNTTTKK249 pKa = 10.54HH250 pKa = 5.62PDD252 pKa = 3.03ILKK255 pKa = 7.88MTGDD259 pKa = 3.85IYY261 pKa = 11.54GNTCWFCVSRR271 pKa = 11.84EE272 pKa = 3.96QMFARR277 pKa = 11.84HH278 pKa = 5.04LWSRR282 pKa = 11.84NGDD285 pKa = 3.55NGDD288 pKa = 4.16PVPHH292 pKa = 6.87TDD294 pKa = 2.97GHH296 pKa = 6.76KK297 pKa = 10.09DD298 pKa = 3.0TSLYY302 pKa = 10.94LSGTGDD308 pKa = 3.97TKK310 pKa = 11.15TMATPVYY317 pKa = 9.7FNTPSGSLLTSEE329 pKa = 4.25TQLFNRR335 pKa = 11.84PFWMQRR341 pKa = 11.84AQGLNNGVCWNNTLFVTVADD361 pKa = 3.92NTRR364 pKa = 11.84GTNLNISVAKK374 pKa = 10.43QEE376 pKa = 4.21GKK378 pKa = 9.97NAEE381 pKa = 4.41SYY383 pKa = 8.1TASNYY388 pKa = 8.41KK389 pKa = 10.07HH390 pKa = 6.24YY391 pKa = 10.73NRR393 pKa = 11.84HH394 pKa = 5.18CEE396 pKa = 3.68IYY398 pKa = 9.47EE399 pKa = 4.11LEE401 pKa = 5.3FILQLATVALTPEE414 pKa = 4.56ALSHH418 pKa = 5.61LHH420 pKa = 7.03AMDD423 pKa = 3.72STILTKK429 pKa = 10.34WNLGFTASNATAVEE443 pKa = 3.98STYY446 pKa = 10.75RR447 pKa = 11.84YY448 pKa = 9.73INSKK452 pKa = 8.25ATKK455 pKa = 9.69CPIDD459 pKa = 3.64IPEE462 pKa = 4.84PEE464 pKa = 4.99PKK466 pKa = 10.1DD467 pKa = 3.44KK468 pKa = 11.46YY469 pKa = 11.25ADD471 pKa = 3.49LTFWNVDD478 pKa = 3.21VSKK481 pKa = 11.0SLSKK485 pKa = 10.78DD486 pKa = 3.23LSAFPLGRR494 pKa = 11.84KK495 pKa = 8.68FLYY498 pKa = 10.02QAGLQNGATRR508 pKa = 11.84SATKK512 pKa = 10.4RR513 pKa = 11.84SATKK517 pKa = 10.06TSTKK521 pKa = 10.31VPTKK525 pKa = 10.1RR526 pKa = 11.84KK527 pKa = 9.53RR528 pKa = 11.84RR529 pKa = 3.71
Molecular weight: 58.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2515 |
171 |
638 |
419.2 |
46.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.203 ± 0.354 | 2.783 ± 0.845 |
6.481 ± 0.559 | 5.129 ± 0.42 |
3.658 ± 0.374 | 7.197 ± 0.373 |
2.584 ± 0.511 | 4.493 ± 0.557 |
5.288 ± 0.889 | 8.986 ± 0.611 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.988 ± 0.303 | 3.817 ± 0.638 |
6.64 ± 0.71 | 4.334 ± 0.88 |
5.01 ± 0.391 | 7.674 ± 0.704 |
7.634 ± 0.908 | 5.686 ± 0.053 |
1.551 ± 0.375 | 2.863 ± 0.215 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
