
African swine fever virus (strain Badajoz 1971 Vero-adapted) (Ba71V) (ASFV)
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Pokkesviricetes; Asfuvirales; Asfarviridae; Asfivirus; African swine fever virus
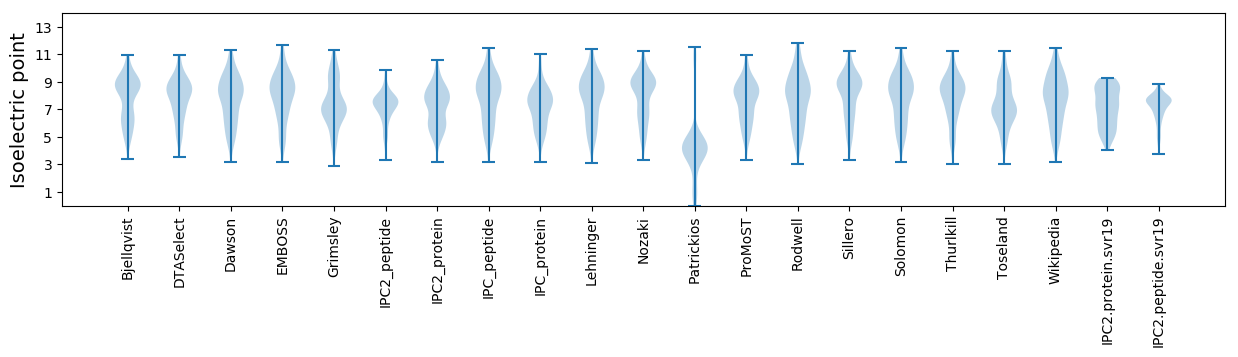
Average proteome isoelectric point is 7.14
Get precalculated fractions of proteins
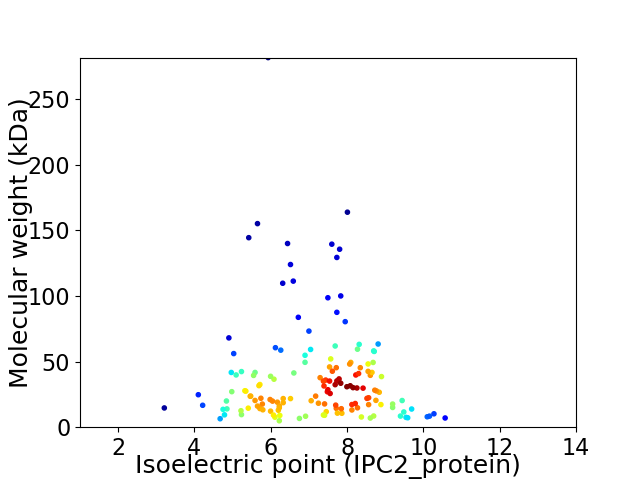
Virtual 2D-PAGE plot for 150 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
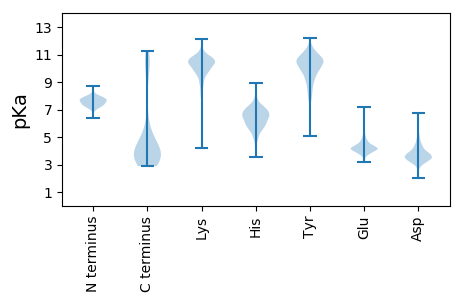
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P32094|MCE_ASFB7 Probable mRNA-capping enzyme OS=African swine fever virus (strain Badajoz 1971 Vero-adapted) OX=10498 GN=Ba71V-101 PE=3 SV=1
MM1 pKa = 7.46VSRR4 pKa = 11.84FLIAEE9 pKa = 4.0YY10 pKa = 10.43RR11 pKa = 11.84HH12 pKa = 6.45LIEE15 pKa = 5.41NPSEE19 pKa = 3.97NFKK22 pKa = 10.92ISVNEE27 pKa = 3.91NNITEE32 pKa = 4.12WDD34 pKa = 3.9VILRR38 pKa = 11.84GPPDD42 pKa = 3.39TLYY45 pKa = 11.18EE46 pKa = 4.56GGLFKK51 pKa = 11.06AKK53 pKa = 10.14VAFPPEE59 pKa = 4.17YY60 pKa = 9.88PYY62 pKa = 11.3APPKK66 pKa = 9.97LTFTSEE72 pKa = 3.38MWHH75 pKa = 6.31PNIYY79 pKa = 9.88PDD81 pKa = 3.75GRR83 pKa = 11.84LCISILHH90 pKa = 6.64GDD92 pKa = 3.56NAEE95 pKa = 4.12EE96 pKa = 5.24QGMTWSPAQKK106 pKa = 9.64IDD108 pKa = 3.88TILLSVISLLNEE120 pKa = 4.11PNPDD124 pKa = 3.04SPANVDD130 pKa = 3.14AAKK133 pKa = 10.28SYY135 pKa = 10.83RR136 pKa = 11.84KK137 pKa = 9.61YY138 pKa = 10.9VYY140 pKa = 10.51KK141 pKa = 10.39EE142 pKa = 3.95DD143 pKa = 3.96LEE145 pKa = 4.52SYY147 pKa = 10.2PMEE150 pKa = 4.05VKK152 pKa = 9.46KK153 pKa = 8.91TVKK156 pKa = 10.25KK157 pKa = 10.92SLDD160 pKa = 3.35EE161 pKa = 4.43CSPEE165 pKa = 4.36DD166 pKa = 3.84IEE168 pKa = 4.43YY169 pKa = 10.53FKK171 pKa = 11.24NAASNVPPIPSDD183 pKa = 3.31AYY185 pKa = 10.13EE186 pKa = 4.47DD187 pKa = 3.69EE188 pKa = 4.66CEE190 pKa = 4.09EE191 pKa = 4.45MEE193 pKa = 5.26DD194 pKa = 3.27DD195 pKa = 4.73TYY197 pKa = 11.61ILTYY201 pKa = 11.0DD202 pKa = 4.06DD203 pKa = 5.65DD204 pKa = 4.52EE205 pKa = 4.86EE206 pKa = 5.45EE207 pKa = 4.44EE208 pKa = 4.36DD209 pKa = 5.05EE210 pKa = 5.3EE211 pKa = 4.71MDD213 pKa = 5.41DD214 pKa = 4.1EE215 pKa = 4.56
MM1 pKa = 7.46VSRR4 pKa = 11.84FLIAEE9 pKa = 4.0YY10 pKa = 10.43RR11 pKa = 11.84HH12 pKa = 6.45LIEE15 pKa = 5.41NPSEE19 pKa = 3.97NFKK22 pKa = 10.92ISVNEE27 pKa = 3.91NNITEE32 pKa = 4.12WDD34 pKa = 3.9VILRR38 pKa = 11.84GPPDD42 pKa = 3.39TLYY45 pKa = 11.18EE46 pKa = 4.56GGLFKK51 pKa = 11.06AKK53 pKa = 10.14VAFPPEE59 pKa = 4.17YY60 pKa = 9.88PYY62 pKa = 11.3APPKK66 pKa = 9.97LTFTSEE72 pKa = 3.38MWHH75 pKa = 6.31PNIYY79 pKa = 9.88PDD81 pKa = 3.75GRR83 pKa = 11.84LCISILHH90 pKa = 6.64GDD92 pKa = 3.56NAEE95 pKa = 4.12EE96 pKa = 5.24QGMTWSPAQKK106 pKa = 9.64IDD108 pKa = 3.88TILLSVISLLNEE120 pKa = 4.11PNPDD124 pKa = 3.04SPANVDD130 pKa = 3.14AAKK133 pKa = 10.28SYY135 pKa = 10.83RR136 pKa = 11.84KK137 pKa = 9.61YY138 pKa = 10.9VYY140 pKa = 10.51KK141 pKa = 10.39EE142 pKa = 3.95DD143 pKa = 3.96LEE145 pKa = 4.52SYY147 pKa = 10.2PMEE150 pKa = 4.05VKK152 pKa = 9.46KK153 pKa = 8.91TVKK156 pKa = 10.25KK157 pKa = 10.92SLDD160 pKa = 3.35EE161 pKa = 4.43CSPEE165 pKa = 4.36DD166 pKa = 3.84IEE168 pKa = 4.43YY169 pKa = 10.53FKK171 pKa = 11.24NAASNVPPIPSDD183 pKa = 3.31AYY185 pKa = 10.13EE186 pKa = 4.47DD187 pKa = 3.69EE188 pKa = 4.66CEE190 pKa = 4.09EE191 pKa = 4.45MEE193 pKa = 5.26DD194 pKa = 3.27DD195 pKa = 4.73TYY197 pKa = 11.61ILTYY201 pKa = 11.0DD202 pKa = 4.06DD203 pKa = 5.65DD204 pKa = 4.52EE205 pKa = 4.86EE206 pKa = 5.45EE207 pKa = 4.44EE208 pKa = 4.36DD209 pKa = 5.05EE210 pKa = 5.3EE211 pKa = 4.71MDD213 pKa = 5.41DD214 pKa = 4.1EE215 pKa = 4.56
Molecular weight: 24.74 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q65162|H962R_ASFB7 Putative helicase C962R OS=African swine fever virus (strain Badajoz 1971 Vero-adapted) OX=10498 GN=Ba71V-071 PE=3 SV=1
MM1 pKa = 7.22NWGSISSGTPGLFVEE16 pKa = 5.55SIRR19 pKa = 11.84NTPSVVKK26 pKa = 10.59INVIFLKK33 pKa = 10.64VISNTAVSVFWRR45 pKa = 11.84DD46 pKa = 2.66RR47 pKa = 11.84RR48 pKa = 11.84IRR50 pKa = 11.84FEE52 pKa = 4.32SDD54 pKa = 2.11WLNSYY59 pKa = 7.17FQKK62 pKa = 11.15
MM1 pKa = 7.22NWGSISSGTPGLFVEE16 pKa = 5.55SIRR19 pKa = 11.84NTPSVVKK26 pKa = 10.59INVIFLKK33 pKa = 10.64VISNTAVSVFWRR45 pKa = 11.84DD46 pKa = 2.66RR47 pKa = 11.84RR48 pKa = 11.84IRR50 pKa = 11.84FEE52 pKa = 4.32SDD54 pKa = 2.11WLNSYY59 pKa = 7.17FQKK62 pKa = 11.15
Molecular weight: 7.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
49966 |
42 |
2475 |
333.1 |
38.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.238 ± 0.175 | 1.987 ± 0.13 |
4.547 ± 0.111 | 6.324 ± 0.146 |
4.795 ± 0.137 | 4.357 ± 0.178 |
2.874 ± 0.119 | 8.314 ± 0.148 |
7.463 ± 0.23 | 10.143 ± 0.178 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.644 ± 0.084 | 6.002 ± 0.16 |
4.353 ± 0.167 | 4.309 ± 0.137 |
4.051 ± 0.166 | 6.136 ± 0.155 |
5.634 ± 0.128 | 5.145 ± 0.121 |
1.061 ± 0.059 | 4.623 ± 0.118 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
