
Leminorella grimontii ATCC 33999 = DSM 5078
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Budviciaceae; Leminorella; Leminorella grimontii
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
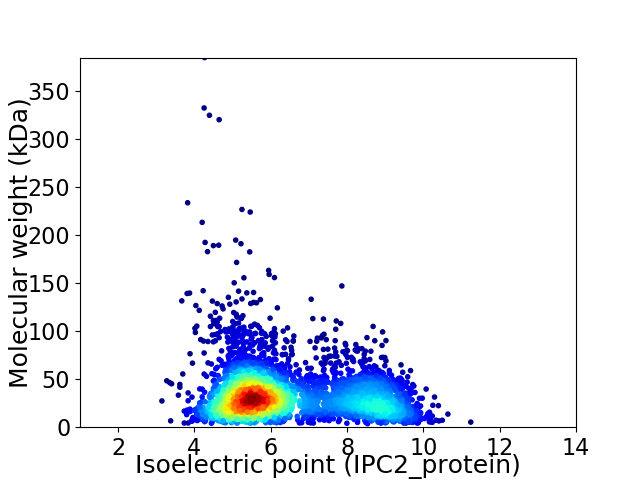
Virtual 2D-PAGE plot for 3756 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A085HGK5|A0A085HGK5_9GAMM Pyruvate dehydrogenase complex transcriptional repressor OS=Leminorella grimontii ATCC 33999 = DSM 5078 OX=1005999 GN=GLGR_2168 PE=4 SV=1
VV1 pKa = 6.23ATDD4 pKa = 3.34AAGNEE9 pKa = 4.63SPASTTFEE17 pKa = 4.2LTVDD21 pKa = 4.25TVAPAAPIITSIVDD35 pKa = 3.46NLSPTTGPIVAGVATNDD52 pKa = 4.01PAPTLNGRR60 pKa = 11.84AEE62 pKa = 4.12AGAIVTIFDD71 pKa = 4.77GEE73 pKa = 4.46DD74 pKa = 3.26NPIGSTTADD83 pKa = 4.47DD84 pKa = 3.8NGDD87 pKa = 2.92WHH89 pKa = 6.3FTPDD93 pKa = 3.41EE94 pKa = 4.15PLGEE98 pKa = 4.49GDD100 pKa = 4.39HH101 pKa = 6.67EE102 pKa = 4.35LTAVATDD109 pKa = 3.25AAGNVGGASPFVAITIDD126 pKa = 3.41TTAPDD131 pKa = 4.01APTGLSVSPSGTRR144 pKa = 11.84VTGSAEE150 pKa = 3.92PGSTVTITDD159 pKa = 3.14ADD161 pKa = 4.05GTVLGSATADD171 pKa = 3.39GTGSFTATITPAQTNGEE188 pKa = 4.25SLLAFAQDD196 pKa = 3.1KK197 pKa = 10.66AGNVGEE203 pKa = 4.24AAGFSASTTGLPDD216 pKa = 3.04VPTIEE221 pKa = 4.32TVTDD225 pKa = 3.48NVSPITGPLEE235 pKa = 4.3NGQSTNEE242 pKa = 4.06TLPLLSGKK250 pKa = 9.43AQAGATVTVYY260 pKa = 11.11DD261 pKa = 4.09KK262 pKa = 11.11GAEE265 pKa = 4.05IGTTTADD272 pKa = 5.48DD273 pKa = 4.33DD274 pKa = 4.72GVWNFTPTVPLTEE287 pKa = 4.42GQHH290 pKa = 5.54VFTATSTNASGTGGFSEE307 pKa = 4.35PRR309 pKa = 11.84SIVVDD314 pKa = 4.88TIAPDD319 pKa = 3.43APTATISADD328 pKa = 2.77GSMISGIAEE337 pKa = 4.21AGSTVTITLANGTTVTTAANDD358 pKa = 3.69SGSYY362 pKa = 10.45SYY364 pKa = 10.44TFQSKK369 pKa = 7.41QTDD372 pKa = 4.01GEE374 pKa = 4.45QLFVTATDD382 pKa = 3.55AAGNPSPRR390 pKa = 11.84TQADD394 pKa = 3.67APDD397 pKa = 4.48LPLSANNNVIEE408 pKa = 4.69LALTSDD414 pKa = 3.96APVTTTQYY422 pKa = 11.35GDD424 pKa = 3.35YY425 pKa = 11.2GFLLVGALGNLASVLGNDD443 pKa = 3.67TAQVTFTVGNGSTADD458 pKa = 3.55VKK460 pKa = 10.73IDD462 pKa = 3.47ADD464 pKa = 3.4ATGIVLSLLSSLEE477 pKa = 4.12LVVQLHH483 pKa = 6.11NSANDD488 pKa = 2.8SWTTVVDD495 pKa = 3.94TQAPRR500 pKa = 11.84WASLLTFGDD509 pKa = 3.64SGISLNLDD517 pKa = 3.01ALAAGTYY524 pKa = 9.79RR525 pKa = 11.84VVSYY529 pKa = 8.38NTSLLATGSYY539 pKa = 10.1TSLNVSVIEE548 pKa = 4.29TGAGTVNGDD557 pKa = 3.54TAHH560 pKa = 6.53SGNVILDD567 pKa = 4.18DD568 pKa = 4.94DD569 pKa = 4.65PVSGSDD575 pKa = 3.36TASGGALVTQVTNAQGTVVPVTADD599 pKa = 3.23GAVIQGTYY607 pKa = 8.79GTLTLYY613 pKa = 10.4PDD615 pKa = 3.86GSYY618 pKa = 10.21TYY620 pKa = 10.7TLFNTSASVIGHH632 pKa = 6.82KK633 pKa = 10.64DD634 pKa = 3.09SFSYY638 pKa = 9.99TLSDD642 pKa = 3.36GTISDD647 pKa = 3.8TASLVISLGEE657 pKa = 3.98GAVTGQAVAVDD668 pKa = 4.16NVASLLFDD676 pKa = 3.84TEE678 pKa = 4.06VATIDD683 pKa = 3.66NGTWSQKK690 pKa = 10.45GYY692 pKa = 8.28TVVNLGLGDD701 pKa = 4.01ALDD704 pKa = 3.86VGVLDD709 pKa = 5.27DD710 pKa = 4.79LSNPIVFNVEE720 pKa = 3.51QGTTRR725 pKa = 11.84SLTLQSEE732 pKa = 4.72VGGIALASKK741 pKa = 10.48FDD743 pKa = 3.76LYY745 pKa = 10.49IYY747 pKa = 10.26KK748 pKa = 10.49FNDD751 pKa = 2.94EE752 pKa = 4.23TQKK755 pKa = 9.85YY756 pKa = 8.31DD757 pKa = 3.09QYY759 pKa = 11.81KK760 pKa = 9.79RR761 pKa = 11.84VEE763 pKa = 4.38DD764 pKa = 3.57WLEE767 pKa = 3.86APLFGGKK774 pKa = 8.91SDD776 pKa = 4.46EE777 pKa = 5.47LDD779 pKa = 3.31LTLDD783 pKa = 3.41GGKK786 pKa = 10.1YY787 pKa = 10.43LFLLDD792 pKa = 3.85TTSGLSVLTGYY803 pKa = 8.74SLKK806 pKa = 10.74VLEE809 pKa = 4.23DD810 pKa = 3.26HH811 pKa = 6.8VYY813 pKa = 10.86SVTSVSASTSGEE825 pKa = 3.99VLDD828 pKa = 5.73NDD830 pKa = 3.57IAPPNTVVSAVNGVSVAASGDD851 pKa = 3.85TVIVGQYY858 pKa = 7.75GTLTINAQGHH868 pKa = 4.24YY869 pKa = 9.48TYY871 pKa = 10.07TLKK874 pKa = 10.97SGVGADD880 pKa = 4.22SISTPDD886 pKa = 3.24SFIYY890 pKa = 10.15TITAPNGDD898 pKa = 4.35RR899 pKa = 11.84DD900 pKa = 4.03TASLNVTPAPRR911 pKa = 11.84GLDD914 pKa = 3.26AVDD917 pKa = 4.25DD918 pKa = 4.27VSAAMTVTSIQDD930 pKa = 3.15TSGYY934 pKa = 10.24SDD936 pKa = 3.37TSVGTASWSSSLGKK950 pKa = 7.52TTGNGSGTFVVDD962 pKa = 3.66EE963 pKa = 4.54GSALKK968 pKa = 10.69DD969 pKa = 2.97IVLTFKK975 pKa = 11.0ASSTLLLGSADD986 pKa = 3.46ISWEE990 pKa = 3.81ILEE993 pKa = 4.51GTTSIRR999 pKa = 11.84SGTTSASMLLGGSPGTVTLSGLEE1022 pKa = 4.1LDD1024 pKa = 4.12AGTYY1028 pKa = 6.98TLKK1031 pKa = 10.1FTGSEE1036 pKa = 4.01TGLVGQITVTPSVSGTLVDD1055 pKa = 4.4LSDD1058 pKa = 3.86YY1059 pKa = 11.15LSTAGHH1065 pKa = 6.03AVSGNIFDD1073 pKa = 4.82GSDD1076 pKa = 2.95SAGAMDD1082 pKa = 3.92QLSSVGTTLSITDD1095 pKa = 3.78ANGVTTTLDD1104 pKa = 3.47PYY1106 pKa = 8.41TTSNATASIHH1116 pKa = 5.2GQYY1119 pKa = 8.93GTLTIGVDD1127 pKa = 3.36GAYY1130 pKa = 10.34SYY1132 pKa = 9.93TLNPAVSLSTITEE1145 pKa = 4.16KK1146 pKa = 10.39EE1147 pKa = 4.27TFNYY1151 pKa = 9.77TLTSSNGQTANATLTVDD1168 pKa = 3.81MAPQFVSSGHH1178 pKa = 6.1NDD1180 pKa = 3.53VITGTAYY1187 pKa = 10.49ADD1189 pKa = 3.31TAIYY1193 pKa = 10.24HH1194 pKa = 6.82LLNAASNTGGNGSDD1208 pKa = 2.9VWSNFSLQQGDD1219 pKa = 4.14KK1220 pKa = 10.52IDD1222 pKa = 3.38IHH1224 pKa = 6.3EE1225 pKa = 5.38LLVGWNGQTSTLGNYY1240 pKa = 10.48LSVTTSGNDD1249 pKa = 3.29TVISIDD1255 pKa = 3.4RR1256 pKa = 11.84DD1257 pKa = 3.58GASGQTYY1264 pKa = 10.27QSTTLITLEE1273 pKa = 4.19NVHH1276 pKa = 5.7TTLDD1280 pKa = 3.55EE1281 pKa = 5.28LIEE1284 pKa = 3.9QHH1286 pKa = 7.08HH1287 pKa = 7.22IITHH1291 pKa = 5.9
VV1 pKa = 6.23ATDD4 pKa = 3.34AAGNEE9 pKa = 4.63SPASTTFEE17 pKa = 4.2LTVDD21 pKa = 4.25TVAPAAPIITSIVDD35 pKa = 3.46NLSPTTGPIVAGVATNDD52 pKa = 4.01PAPTLNGRR60 pKa = 11.84AEE62 pKa = 4.12AGAIVTIFDD71 pKa = 4.77GEE73 pKa = 4.46DD74 pKa = 3.26NPIGSTTADD83 pKa = 4.47DD84 pKa = 3.8NGDD87 pKa = 2.92WHH89 pKa = 6.3FTPDD93 pKa = 3.41EE94 pKa = 4.15PLGEE98 pKa = 4.49GDD100 pKa = 4.39HH101 pKa = 6.67EE102 pKa = 4.35LTAVATDD109 pKa = 3.25AAGNVGGASPFVAITIDD126 pKa = 3.41TTAPDD131 pKa = 4.01APTGLSVSPSGTRR144 pKa = 11.84VTGSAEE150 pKa = 3.92PGSTVTITDD159 pKa = 3.14ADD161 pKa = 4.05GTVLGSATADD171 pKa = 3.39GTGSFTATITPAQTNGEE188 pKa = 4.25SLLAFAQDD196 pKa = 3.1KK197 pKa = 10.66AGNVGEE203 pKa = 4.24AAGFSASTTGLPDD216 pKa = 3.04VPTIEE221 pKa = 4.32TVTDD225 pKa = 3.48NVSPITGPLEE235 pKa = 4.3NGQSTNEE242 pKa = 4.06TLPLLSGKK250 pKa = 9.43AQAGATVTVYY260 pKa = 11.11DD261 pKa = 4.09KK262 pKa = 11.11GAEE265 pKa = 4.05IGTTTADD272 pKa = 5.48DD273 pKa = 4.33DD274 pKa = 4.72GVWNFTPTVPLTEE287 pKa = 4.42GQHH290 pKa = 5.54VFTATSTNASGTGGFSEE307 pKa = 4.35PRR309 pKa = 11.84SIVVDD314 pKa = 4.88TIAPDD319 pKa = 3.43APTATISADD328 pKa = 2.77GSMISGIAEE337 pKa = 4.21AGSTVTITLANGTTVTTAANDD358 pKa = 3.69SGSYY362 pKa = 10.45SYY364 pKa = 10.44TFQSKK369 pKa = 7.41QTDD372 pKa = 4.01GEE374 pKa = 4.45QLFVTATDD382 pKa = 3.55AAGNPSPRR390 pKa = 11.84TQADD394 pKa = 3.67APDD397 pKa = 4.48LPLSANNNVIEE408 pKa = 4.69LALTSDD414 pKa = 3.96APVTTTQYY422 pKa = 11.35GDD424 pKa = 3.35YY425 pKa = 11.2GFLLVGALGNLASVLGNDD443 pKa = 3.67TAQVTFTVGNGSTADD458 pKa = 3.55VKK460 pKa = 10.73IDD462 pKa = 3.47ADD464 pKa = 3.4ATGIVLSLLSSLEE477 pKa = 4.12LVVQLHH483 pKa = 6.11NSANDD488 pKa = 2.8SWTTVVDD495 pKa = 3.94TQAPRR500 pKa = 11.84WASLLTFGDD509 pKa = 3.64SGISLNLDD517 pKa = 3.01ALAAGTYY524 pKa = 9.79RR525 pKa = 11.84VVSYY529 pKa = 8.38NTSLLATGSYY539 pKa = 10.1TSLNVSVIEE548 pKa = 4.29TGAGTVNGDD557 pKa = 3.54TAHH560 pKa = 6.53SGNVILDD567 pKa = 4.18DD568 pKa = 4.94DD569 pKa = 4.65PVSGSDD575 pKa = 3.36TASGGALVTQVTNAQGTVVPVTADD599 pKa = 3.23GAVIQGTYY607 pKa = 8.79GTLTLYY613 pKa = 10.4PDD615 pKa = 3.86GSYY618 pKa = 10.21TYY620 pKa = 10.7TLFNTSASVIGHH632 pKa = 6.82KK633 pKa = 10.64DD634 pKa = 3.09SFSYY638 pKa = 9.99TLSDD642 pKa = 3.36GTISDD647 pKa = 3.8TASLVISLGEE657 pKa = 3.98GAVTGQAVAVDD668 pKa = 4.16NVASLLFDD676 pKa = 3.84TEE678 pKa = 4.06VATIDD683 pKa = 3.66NGTWSQKK690 pKa = 10.45GYY692 pKa = 8.28TVVNLGLGDD701 pKa = 4.01ALDD704 pKa = 3.86VGVLDD709 pKa = 5.27DD710 pKa = 4.79LSNPIVFNVEE720 pKa = 3.51QGTTRR725 pKa = 11.84SLTLQSEE732 pKa = 4.72VGGIALASKK741 pKa = 10.48FDD743 pKa = 3.76LYY745 pKa = 10.49IYY747 pKa = 10.26KK748 pKa = 10.49FNDD751 pKa = 2.94EE752 pKa = 4.23TQKK755 pKa = 9.85YY756 pKa = 8.31DD757 pKa = 3.09QYY759 pKa = 11.81KK760 pKa = 9.79RR761 pKa = 11.84VEE763 pKa = 4.38DD764 pKa = 3.57WLEE767 pKa = 3.86APLFGGKK774 pKa = 8.91SDD776 pKa = 4.46EE777 pKa = 5.47LDD779 pKa = 3.31LTLDD783 pKa = 3.41GGKK786 pKa = 10.1YY787 pKa = 10.43LFLLDD792 pKa = 3.85TTSGLSVLTGYY803 pKa = 8.74SLKK806 pKa = 10.74VLEE809 pKa = 4.23DD810 pKa = 3.26HH811 pKa = 6.8VYY813 pKa = 10.86SVTSVSASTSGEE825 pKa = 3.99VLDD828 pKa = 5.73NDD830 pKa = 3.57IAPPNTVVSAVNGVSVAASGDD851 pKa = 3.85TVIVGQYY858 pKa = 7.75GTLTINAQGHH868 pKa = 4.24YY869 pKa = 9.48TYY871 pKa = 10.07TLKK874 pKa = 10.97SGVGADD880 pKa = 4.22SISTPDD886 pKa = 3.24SFIYY890 pKa = 10.15TITAPNGDD898 pKa = 4.35RR899 pKa = 11.84DD900 pKa = 4.03TASLNVTPAPRR911 pKa = 11.84GLDD914 pKa = 3.26AVDD917 pKa = 4.25DD918 pKa = 4.27VSAAMTVTSIQDD930 pKa = 3.15TSGYY934 pKa = 10.24SDD936 pKa = 3.37TSVGTASWSSSLGKK950 pKa = 7.52TTGNGSGTFVVDD962 pKa = 3.66EE963 pKa = 4.54GSALKK968 pKa = 10.69DD969 pKa = 2.97IVLTFKK975 pKa = 11.0ASSTLLLGSADD986 pKa = 3.46ISWEE990 pKa = 3.81ILEE993 pKa = 4.51GTTSIRR999 pKa = 11.84SGTTSASMLLGGSPGTVTLSGLEE1022 pKa = 4.1LDD1024 pKa = 4.12AGTYY1028 pKa = 6.98TLKK1031 pKa = 10.1FTGSEE1036 pKa = 4.01TGLVGQITVTPSVSGTLVDD1055 pKa = 4.4LSDD1058 pKa = 3.86YY1059 pKa = 11.15LSTAGHH1065 pKa = 6.03AVSGNIFDD1073 pKa = 4.82GSDD1076 pKa = 2.95SAGAMDD1082 pKa = 3.92QLSSVGTTLSITDD1095 pKa = 3.78ANGVTTTLDD1104 pKa = 3.47PYY1106 pKa = 8.41TTSNATASIHH1116 pKa = 5.2GQYY1119 pKa = 8.93GTLTIGVDD1127 pKa = 3.36GAYY1130 pKa = 10.34SYY1132 pKa = 9.93TLNPAVSLSTITEE1145 pKa = 4.16KK1146 pKa = 10.39EE1147 pKa = 4.27TFNYY1151 pKa = 9.77TLTSSNGQTANATLTVDD1168 pKa = 3.81MAPQFVSSGHH1178 pKa = 6.1NDD1180 pKa = 3.53VITGTAYY1187 pKa = 10.49ADD1189 pKa = 3.31TAIYY1193 pKa = 10.24HH1194 pKa = 6.82LLNAASNTGGNGSDD1208 pKa = 2.9VWSNFSLQQGDD1219 pKa = 4.14KK1220 pKa = 10.52IDD1222 pKa = 3.38IHH1224 pKa = 6.3EE1225 pKa = 5.38LLVGWNGQTSTLGNYY1240 pKa = 10.48LSVTTSGNDD1249 pKa = 3.29TVISIDD1255 pKa = 3.4RR1256 pKa = 11.84DD1257 pKa = 3.58GASGQTYY1264 pKa = 10.27QSTTLITLEE1273 pKa = 4.19NVHH1276 pKa = 5.7TTLDD1280 pKa = 3.55EE1281 pKa = 5.28LIEE1284 pKa = 3.9QHH1286 pKa = 7.08HH1287 pKa = 7.22IITHH1291 pKa = 5.9
Molecular weight: 131.35 kDa
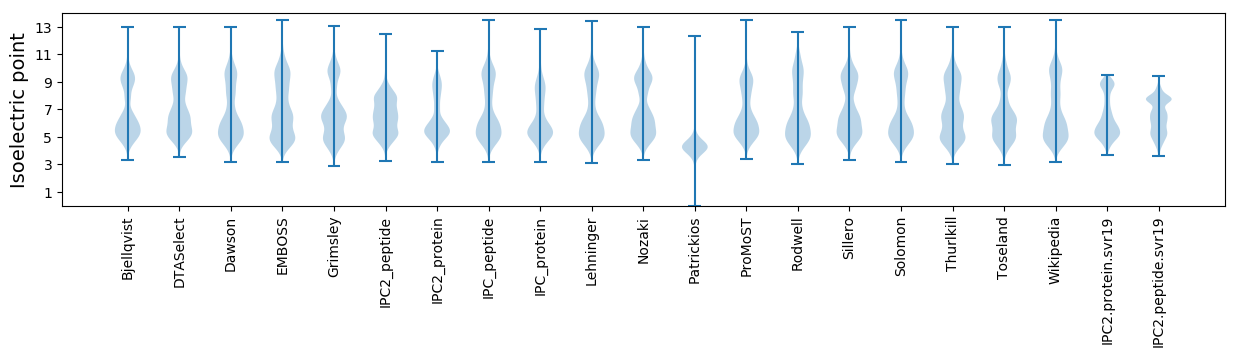
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A085HBX3|A0A085HBX3_9GAMM DNA gyrase subunit B OS=Leminorella grimontii ATCC 33999 = DSM 5078 OX=1005999 GN=gyrB PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.62RR12 pKa = 11.84NRR14 pKa = 11.84AHH16 pKa = 7.59GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.16GRR39 pKa = 11.84SRR41 pKa = 11.84LTVAGG46 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.62RR12 pKa = 11.84NRR14 pKa = 11.84AHH16 pKa = 7.59GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.16GRR39 pKa = 11.84SRR41 pKa = 11.84LTVAGG46 pKa = 3.91
Molecular weight: 5.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1211632 |
37 |
3852 |
322.6 |
35.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.623 ± 0.047 | 1.079 ± 0.016 |
5.344 ± 0.033 | 5.778 ± 0.042 |
3.799 ± 0.032 | 7.947 ± 0.067 |
1.971 ± 0.018 | 5.838 ± 0.037 |
4.478 ± 0.036 | 10.458 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.643 ± 0.023 | 3.871 ± 0.043 |
4.22 ± 0.035 | 4.013 ± 0.032 |
5.512 ± 0.042 | 6.535 ± 0.041 |
5.346 ± 0.042 | 7.176 ± 0.029 |
1.335 ± 0.017 | 3.035 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
