
Botryosphaeria dothidea victorivirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Victorivirus; unclassified Victorivirus
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
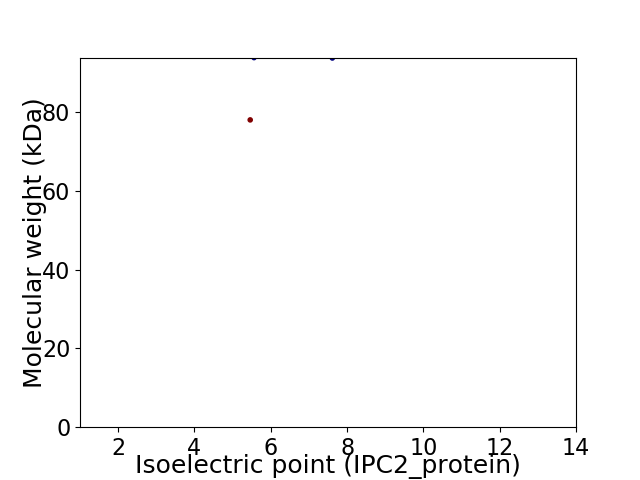
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A089H6F5|A0A089H6F5_9VIRU Coat protein OS=Botryosphaeria dothidea victorivirus 1 OX=1547580 GN=CP PE=4 SV=1
MM1 pKa = 7.23EE2 pKa = 5.08VEE4 pKa = 4.34TQFRR8 pKa = 11.84PSFLAGILLRR18 pKa = 11.84PTGGRR23 pKa = 11.84LAAEE27 pKa = 4.22AGGRR31 pKa = 11.84EE32 pKa = 4.27YY33 pKa = 11.31ASALNSTMTVRR44 pKa = 11.84GITTTRR50 pKa = 11.84SSVVRR55 pKa = 11.84YY56 pKa = 7.26HH57 pKa = 6.7TSPTQGEE64 pKa = 4.41LRR66 pKa = 11.84DD67 pKa = 4.13KK68 pKa = 11.28LDD70 pKa = 4.21LAPEE74 pKa = 4.58GARR77 pKa = 11.84RR78 pKa = 11.84LVAEE82 pKa = 4.21YY83 pKa = 8.23QTSGEE88 pKa = 4.22MVAHH92 pKa = 6.68FSNLARR98 pKa = 11.84KK99 pKa = 8.68YY100 pKa = 10.88VNFSANFEE108 pKa = 4.13ASNLAGLVEE117 pKa = 5.09RR118 pKa = 11.84ISLGLSLAAWHH129 pKa = 7.18DD130 pKa = 3.9NVTAFSLRR138 pKa = 11.84GGRR141 pKa = 11.84EE142 pKa = 3.42PDD144 pKa = 3.08IAALGATTTPVNSVADD160 pKa = 3.66CVFIPRR166 pKa = 11.84VVNTVITGDD175 pKa = 3.26IFSVLVAAATGCGAAVATDD194 pKa = 3.53MLGVNDD200 pKa = 4.69NGVALLNDD208 pKa = 2.96VDD210 pKa = 4.01ARR212 pKa = 11.84RR213 pKa = 11.84FPPAAVEE220 pKa = 3.92ALRR223 pKa = 11.84LLGANMNDD231 pKa = 3.42AGQGALFALALTRR244 pKa = 11.84GLHH247 pKa = 6.16KK248 pKa = 10.41GTVVQSTTRR257 pKa = 11.84GGALTRR263 pKa = 11.84AFLIAGTFSVPFGGINPTPEE283 pKa = 4.35PYY285 pKa = 10.4AALPALNSSDD295 pKa = 3.18GVAVGGYY302 pKa = 10.25VDD304 pKa = 4.68GLCLATAAAVAHH316 pKa = 6.91CDD318 pKa = 3.07PGTTIEE324 pKa = 4.29GLWYY328 pKa = 6.58PTTYY332 pKa = 8.9VTPRR336 pKa = 11.84HH337 pKa = 6.13DD338 pKa = 3.77AFGVAGSVPTADD350 pKa = 4.15PLNGASLWPQIISEE364 pKa = 4.21SGRR367 pKa = 11.84FLGEE371 pKa = 3.78YY372 pKa = 10.13VSALGRR378 pKa = 11.84LFSAEE383 pKa = 4.48DD384 pKa = 3.73GLGLANRR391 pKa = 11.84AACTMAHH398 pKa = 7.12LALVDD403 pKa = 4.66DD404 pKa = 5.18WGCDD408 pKa = 3.55DD409 pKa = 4.49ATVAPYY415 pKa = 9.79FWIEE419 pKa = 3.88PTSILPQNWLGSAAEE434 pKa = 4.11RR435 pKa = 11.84SMSGALVTPGGEE447 pKa = 4.07VTLPAVEE454 pKa = 4.62GVQPVGDD461 pKa = 4.09STGPWAGYY469 pKa = 8.54VLEE472 pKa = 4.74LSSARR477 pKa = 11.84RR478 pKa = 11.84NPLFIHH484 pKa = 6.84FGGQMSEE491 pKa = 4.02HH492 pKa = 7.06LGTIVPRR499 pKa = 11.84QLDD502 pKa = 4.0VEE504 pKa = 4.79TVVHH508 pKa = 6.12PKK510 pKa = 10.16LRR512 pKa = 11.84AEE514 pKa = 3.99QAALRR519 pKa = 11.84DD520 pKa = 3.86RR521 pKa = 11.84LQASLGLDD529 pKa = 3.29QYY531 pKa = 10.81VHH533 pKa = 6.89LPEE536 pKa = 5.11RR537 pKa = 11.84MPIPSPDD544 pKa = 3.14NFMHH548 pKa = 6.05VGCRR552 pKa = 11.84MGVEE556 pKa = 4.5VVHH559 pKa = 5.81EE560 pKa = 3.98QFVNYY565 pKa = 10.07RR566 pKa = 11.84MYY568 pKa = 11.2PSMLPAAHH576 pKa = 6.85EE577 pKa = 4.46FAHH580 pKa = 5.6STVTLSVTKK589 pKa = 10.3PFGVPAGAAPEE600 pKa = 4.19IEE602 pKa = 4.79CGFSHH607 pKa = 7.17EE608 pKa = 4.23PTRR611 pKa = 11.84TALMLSQTSHH621 pKa = 6.21NLRR624 pKa = 11.84GPSSVAFTGMLISTSAPAAPRR645 pKa = 11.84YY646 pKa = 8.27TSGQGRR652 pKa = 11.84SRR654 pKa = 11.84RR655 pKa = 11.84VALGDD660 pKa = 3.32EE661 pKa = 4.15RR662 pKa = 11.84HH663 pKa = 5.74EE664 pKa = 4.35VGSAGVDD671 pKa = 3.56HH672 pKa = 6.86NPSPPVPVGNRR683 pKa = 11.84LGVTTHH689 pKa = 6.78HH690 pKa = 7.13DD691 pKa = 3.63AGRR694 pKa = 11.84GPAPIRR700 pKa = 11.84RR701 pKa = 11.84ADD703 pKa = 3.44QVRR706 pKa = 11.84GGGGGAPQRR715 pKa = 11.84RR716 pKa = 11.84RR717 pKa = 11.84DD718 pKa = 3.78DD719 pKa = 3.88GQAQDD724 pKa = 4.45PVAPPPAHH732 pKa = 6.94GEE734 pKa = 3.9PAPPEE739 pKa = 3.87VDD741 pKa = 3.35EE742 pKa = 4.52PP743 pKa = 4.08
MM1 pKa = 7.23EE2 pKa = 5.08VEE4 pKa = 4.34TQFRR8 pKa = 11.84PSFLAGILLRR18 pKa = 11.84PTGGRR23 pKa = 11.84LAAEE27 pKa = 4.22AGGRR31 pKa = 11.84EE32 pKa = 4.27YY33 pKa = 11.31ASALNSTMTVRR44 pKa = 11.84GITTTRR50 pKa = 11.84SSVVRR55 pKa = 11.84YY56 pKa = 7.26HH57 pKa = 6.7TSPTQGEE64 pKa = 4.41LRR66 pKa = 11.84DD67 pKa = 4.13KK68 pKa = 11.28LDD70 pKa = 4.21LAPEE74 pKa = 4.58GARR77 pKa = 11.84RR78 pKa = 11.84LVAEE82 pKa = 4.21YY83 pKa = 8.23QTSGEE88 pKa = 4.22MVAHH92 pKa = 6.68FSNLARR98 pKa = 11.84KK99 pKa = 8.68YY100 pKa = 10.88VNFSANFEE108 pKa = 4.13ASNLAGLVEE117 pKa = 5.09RR118 pKa = 11.84ISLGLSLAAWHH129 pKa = 7.18DD130 pKa = 3.9NVTAFSLRR138 pKa = 11.84GGRR141 pKa = 11.84EE142 pKa = 3.42PDD144 pKa = 3.08IAALGATTTPVNSVADD160 pKa = 3.66CVFIPRR166 pKa = 11.84VVNTVITGDD175 pKa = 3.26IFSVLVAAATGCGAAVATDD194 pKa = 3.53MLGVNDD200 pKa = 4.69NGVALLNDD208 pKa = 2.96VDD210 pKa = 4.01ARR212 pKa = 11.84RR213 pKa = 11.84FPPAAVEE220 pKa = 3.92ALRR223 pKa = 11.84LLGANMNDD231 pKa = 3.42AGQGALFALALTRR244 pKa = 11.84GLHH247 pKa = 6.16KK248 pKa = 10.41GTVVQSTTRR257 pKa = 11.84GGALTRR263 pKa = 11.84AFLIAGTFSVPFGGINPTPEE283 pKa = 4.35PYY285 pKa = 10.4AALPALNSSDD295 pKa = 3.18GVAVGGYY302 pKa = 10.25VDD304 pKa = 4.68GLCLATAAAVAHH316 pKa = 6.91CDD318 pKa = 3.07PGTTIEE324 pKa = 4.29GLWYY328 pKa = 6.58PTTYY332 pKa = 8.9VTPRR336 pKa = 11.84HH337 pKa = 6.13DD338 pKa = 3.77AFGVAGSVPTADD350 pKa = 4.15PLNGASLWPQIISEE364 pKa = 4.21SGRR367 pKa = 11.84FLGEE371 pKa = 3.78YY372 pKa = 10.13VSALGRR378 pKa = 11.84LFSAEE383 pKa = 4.48DD384 pKa = 3.73GLGLANRR391 pKa = 11.84AACTMAHH398 pKa = 7.12LALVDD403 pKa = 4.66DD404 pKa = 5.18WGCDD408 pKa = 3.55DD409 pKa = 4.49ATVAPYY415 pKa = 9.79FWIEE419 pKa = 3.88PTSILPQNWLGSAAEE434 pKa = 4.11RR435 pKa = 11.84SMSGALVTPGGEE447 pKa = 4.07VTLPAVEE454 pKa = 4.62GVQPVGDD461 pKa = 4.09STGPWAGYY469 pKa = 8.54VLEE472 pKa = 4.74LSSARR477 pKa = 11.84RR478 pKa = 11.84NPLFIHH484 pKa = 6.84FGGQMSEE491 pKa = 4.02HH492 pKa = 7.06LGTIVPRR499 pKa = 11.84QLDD502 pKa = 4.0VEE504 pKa = 4.79TVVHH508 pKa = 6.12PKK510 pKa = 10.16LRR512 pKa = 11.84AEE514 pKa = 3.99QAALRR519 pKa = 11.84DD520 pKa = 3.86RR521 pKa = 11.84LQASLGLDD529 pKa = 3.29QYY531 pKa = 10.81VHH533 pKa = 6.89LPEE536 pKa = 5.11RR537 pKa = 11.84MPIPSPDD544 pKa = 3.14NFMHH548 pKa = 6.05VGCRR552 pKa = 11.84MGVEE556 pKa = 4.5VVHH559 pKa = 5.81EE560 pKa = 3.98QFVNYY565 pKa = 10.07RR566 pKa = 11.84MYY568 pKa = 11.2PSMLPAAHH576 pKa = 6.85EE577 pKa = 4.46FAHH580 pKa = 5.6STVTLSVTKK589 pKa = 10.3PFGVPAGAAPEE600 pKa = 4.19IEE602 pKa = 4.79CGFSHH607 pKa = 7.17EE608 pKa = 4.23PTRR611 pKa = 11.84TALMLSQTSHH621 pKa = 6.21NLRR624 pKa = 11.84GPSSVAFTGMLISTSAPAAPRR645 pKa = 11.84YY646 pKa = 8.27TSGQGRR652 pKa = 11.84SRR654 pKa = 11.84RR655 pKa = 11.84VALGDD660 pKa = 3.32EE661 pKa = 4.15RR662 pKa = 11.84HH663 pKa = 5.74EE664 pKa = 4.35VGSAGVDD671 pKa = 3.56HH672 pKa = 6.86NPSPPVPVGNRR683 pKa = 11.84LGVTTHH689 pKa = 6.78HH690 pKa = 7.13DD691 pKa = 3.63AGRR694 pKa = 11.84GPAPIRR700 pKa = 11.84RR701 pKa = 11.84ADD703 pKa = 3.44QVRR706 pKa = 11.84GGGGGAPQRR715 pKa = 11.84RR716 pKa = 11.84RR717 pKa = 11.84DD718 pKa = 3.78DD719 pKa = 3.88GQAQDD724 pKa = 4.45PVAPPPAHH732 pKa = 6.94GEE734 pKa = 3.9PAPPEE739 pKa = 3.87VDD741 pKa = 3.35EE742 pKa = 4.52PP743 pKa = 4.08
Molecular weight: 78.06 kDa
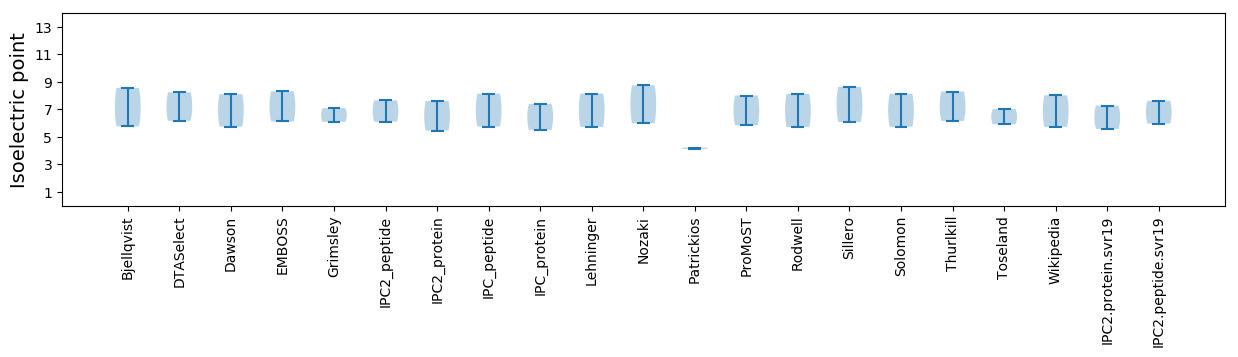
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A089H6F5|A0A089H6F5_9VIRU Coat protein OS=Botryosphaeria dothidea victorivirus 1 OX=1547580 GN=CP PE=4 SV=1
MM1 pKa = 7.17QDD3 pKa = 2.92AVAVRR8 pKa = 11.84AAEE11 pKa = 3.98VGPLGVEE18 pKa = 4.18LLRR21 pKa = 11.84VIDD24 pKa = 4.83TFAAWTRR31 pKa = 11.84THH33 pKa = 7.2FPRR36 pKa = 11.84EE37 pKa = 3.95LVAGMSRR44 pKa = 11.84LQVQRR49 pKa = 11.84QQLDD53 pKa = 3.7SIHH56 pKa = 7.05PLLAPAIATLLLDD69 pKa = 3.71YY70 pKa = 10.38PLQLEE75 pKa = 4.37LDD77 pKa = 3.85HH78 pKa = 6.5GVVMRR83 pKa = 11.84LVDD86 pKa = 4.16LALPRR91 pKa = 11.84IDD93 pKa = 5.33AEE95 pKa = 4.04PAEE98 pKa = 4.25PAKK101 pKa = 10.83NQALRR106 pKa = 11.84QHH108 pKa = 6.35VKK110 pKa = 10.43RR111 pKa = 11.84NSPRR115 pKa = 11.84HH116 pKa = 4.56NQYY119 pKa = 9.6MEE121 pKa = 3.97LASAIRR127 pKa = 11.84HH128 pKa = 5.7DD129 pKa = 3.79KK130 pKa = 10.4PFRR133 pKa = 11.84LACFPEE139 pKa = 4.76KK140 pKa = 10.43KK141 pKa = 10.18LQAATAKK148 pKa = 10.53KK149 pKa = 9.93NFSTRR154 pKa = 11.84RR155 pKa = 11.84LLDD158 pKa = 3.48SVSRR162 pKa = 11.84EE163 pKa = 3.85LGASFLGWLVAHH175 pKa = 6.79CACKK179 pKa = 9.98VTDD182 pKa = 4.72DD183 pKa = 3.96QFQMIIIFGLTLSTRR198 pKa = 11.84LGRR201 pKa = 11.84HH202 pKa = 5.71AYY204 pKa = 10.05LYY206 pKa = 10.43ALSMVTNPSHH216 pKa = 6.85AKK218 pKa = 9.6SLNVVLKK225 pKa = 10.88GLGANASTPGCWFVEE240 pKa = 4.5GQGLLGRR247 pKa = 11.84GVGDD251 pKa = 3.31VDD253 pKa = 3.82WNSEE257 pKa = 3.42IPYY260 pKa = 10.13RR261 pKa = 11.84CDD263 pKa = 3.31PDD265 pKa = 3.71LVRR268 pKa = 11.84EE269 pKa = 4.05KK270 pKa = 10.05TINVDD275 pKa = 3.22PEE277 pKa = 4.29TIRR280 pKa = 11.84PHH282 pKa = 5.72IRR284 pKa = 11.84AILEE288 pKa = 4.01RR289 pKa = 11.84EE290 pKa = 4.01LVNRR294 pKa = 11.84NDD296 pKa = 3.94LPDD299 pKa = 4.47MEE301 pKa = 5.82DD302 pKa = 2.98FWTSRR307 pKa = 11.84WLWCVNGSQTSDD319 pKa = 2.83SDD321 pKa = 3.61KK322 pKa = 11.6ALGLDD327 pKa = 3.92LKK329 pKa = 9.88TLQTHH334 pKa = 5.07RR335 pKa = 11.84RR336 pKa = 11.84RR337 pKa = 11.84YY338 pKa = 9.75RR339 pKa = 11.84RR340 pKa = 11.84MAAEE344 pKa = 4.16ALVDD348 pKa = 4.0NPIPDD353 pKa = 3.79WDD355 pKa = 3.53GTTYY359 pKa = 10.89VSASSKK365 pKa = 11.11LEE367 pKa = 3.85VGKK370 pKa = 8.55TRR372 pKa = 11.84AIFACDD378 pKa = 3.1TRR380 pKa = 11.84SYY382 pKa = 11.04FAFSYY387 pKa = 10.91LLNQVQKK394 pKa = 9.94DD395 pKa = 3.44WRR397 pKa = 11.84NEE399 pKa = 3.78RR400 pKa = 11.84VLLDD404 pKa = 4.04PGNGGTVAMGARR416 pKa = 11.84LRR418 pKa = 11.84NAQKK422 pKa = 10.89GGGFNLMLDD431 pKa = 3.58YY432 pKa = 11.47DD433 pKa = 4.75DD434 pKa = 6.31FNSQHH439 pKa = 5.4STEE442 pKa = 3.96TMKK445 pKa = 10.63IVFEE449 pKa = 4.43EE450 pKa = 4.25TCKK453 pKa = 10.39IYY455 pKa = 10.56NAPAWYY461 pKa = 8.68TDD463 pKa = 3.74KK464 pKa = 11.22LCSSFDD470 pKa = 3.18KK471 pKa = 10.65MYY473 pKa = 10.49IRR475 pKa = 11.84QKK477 pKa = 10.92DD478 pKa = 3.84GLRR481 pKa = 11.84HH482 pKa = 4.58VAGTLMSGHH491 pKa = 7.15RR492 pKa = 11.84GTTFINSVLNAAYY505 pKa = 9.78LRR507 pKa = 11.84CGVGSGWFDD516 pKa = 3.12HH517 pKa = 6.82ALSLHH522 pKa = 6.26TGDD525 pKa = 5.04DD526 pKa = 3.7VYY528 pKa = 11.08IRR530 pKa = 11.84SNSRR534 pKa = 11.84SEE536 pKa = 3.82VSNILTRR543 pKa = 11.84AAEE546 pKa = 4.58FGCRR550 pKa = 11.84MNPTKK555 pKa = 10.6QSIGVKK561 pKa = 8.6NCEE564 pKa = 4.22FLRR567 pKa = 11.84CAYY570 pKa = 10.18NPYY573 pKa = 9.91YY574 pKa = 10.63AVGYY578 pKa = 9.26LCRR581 pKa = 11.84TVGTLVNGNWSGDD594 pKa = 3.66TPLTPHH600 pKa = 6.86EE601 pKa = 4.92ALTSLLTSLRR611 pKa = 11.84SLYY614 pKa = 10.3NRR616 pKa = 11.84SLGGGLGRR624 pKa = 11.84YY625 pKa = 7.71LASAIRR631 pKa = 11.84FRR633 pKa = 11.84TDD635 pKa = 3.71GISRR639 pKa = 11.84KK640 pKa = 9.28NLIGLLDD647 pKa = 3.55GRR649 pKa = 11.84IAIEE653 pKa = 4.82GGPCFNADD661 pKa = 2.99AKK663 pKa = 10.83IRR665 pKa = 11.84TCALKK670 pKa = 9.19NTIKK674 pKa = 10.63PSPLVDD680 pKa = 4.2KK681 pKa = 11.19NWDD684 pKa = 3.14AYY686 pKa = 9.38ATRR689 pKa = 11.84DD690 pKa = 3.65YY691 pKa = 10.82LVCHH695 pKa = 5.61TTDD698 pKa = 3.21LEE700 pKa = 4.55YY701 pKa = 11.12YY702 pKa = 10.25ALQNCGVSPTPLMVTTSYY720 pKa = 11.41EE721 pKa = 3.68KK722 pKa = 10.71GLNRR726 pKa = 11.84EE727 pKa = 4.14SEE729 pKa = 4.28EE730 pKa = 4.0VKK732 pKa = 10.42PLKK735 pKa = 9.77FCNMRR740 pKa = 11.84VSTARR745 pKa = 11.84GFATVSEE752 pKa = 4.18LLKK755 pKa = 10.59VDD757 pKa = 3.6TEE759 pKa = 4.71GGVLAHH765 pKa = 6.48YY766 pKa = 8.56PLLMLIKK773 pKa = 10.5DD774 pKa = 3.97SLSPSLIRR782 pKa = 11.84EE783 pKa = 4.07LCEE786 pKa = 3.84ADD788 pKa = 4.67GYY790 pKa = 9.29TPPSEE795 pKa = 4.53NIRR798 pKa = 11.84AYY800 pKa = 10.65AFGEE804 pKa = 4.01DD805 pKa = 3.86FRR807 pKa = 11.84THH809 pKa = 6.64NIIGRR814 pKa = 11.84LPYY817 pKa = 10.3SDD819 pKa = 4.93AAALQGKK826 pKa = 6.27TSCDD830 pKa = 3.59NIVADD835 pKa = 3.82YY836 pKa = 10.71KK837 pKa = 10.88IYY839 pKa = 10.97LL840 pKa = 3.86
MM1 pKa = 7.17QDD3 pKa = 2.92AVAVRR8 pKa = 11.84AAEE11 pKa = 3.98VGPLGVEE18 pKa = 4.18LLRR21 pKa = 11.84VIDD24 pKa = 4.83TFAAWTRR31 pKa = 11.84THH33 pKa = 7.2FPRR36 pKa = 11.84EE37 pKa = 3.95LVAGMSRR44 pKa = 11.84LQVQRR49 pKa = 11.84QQLDD53 pKa = 3.7SIHH56 pKa = 7.05PLLAPAIATLLLDD69 pKa = 3.71YY70 pKa = 10.38PLQLEE75 pKa = 4.37LDD77 pKa = 3.85HH78 pKa = 6.5GVVMRR83 pKa = 11.84LVDD86 pKa = 4.16LALPRR91 pKa = 11.84IDD93 pKa = 5.33AEE95 pKa = 4.04PAEE98 pKa = 4.25PAKK101 pKa = 10.83NQALRR106 pKa = 11.84QHH108 pKa = 6.35VKK110 pKa = 10.43RR111 pKa = 11.84NSPRR115 pKa = 11.84HH116 pKa = 4.56NQYY119 pKa = 9.6MEE121 pKa = 3.97LASAIRR127 pKa = 11.84HH128 pKa = 5.7DD129 pKa = 3.79KK130 pKa = 10.4PFRR133 pKa = 11.84LACFPEE139 pKa = 4.76KK140 pKa = 10.43KK141 pKa = 10.18LQAATAKK148 pKa = 10.53KK149 pKa = 9.93NFSTRR154 pKa = 11.84RR155 pKa = 11.84LLDD158 pKa = 3.48SVSRR162 pKa = 11.84EE163 pKa = 3.85LGASFLGWLVAHH175 pKa = 6.79CACKK179 pKa = 9.98VTDD182 pKa = 4.72DD183 pKa = 3.96QFQMIIIFGLTLSTRR198 pKa = 11.84LGRR201 pKa = 11.84HH202 pKa = 5.71AYY204 pKa = 10.05LYY206 pKa = 10.43ALSMVTNPSHH216 pKa = 6.85AKK218 pKa = 9.6SLNVVLKK225 pKa = 10.88GLGANASTPGCWFVEE240 pKa = 4.5GQGLLGRR247 pKa = 11.84GVGDD251 pKa = 3.31VDD253 pKa = 3.82WNSEE257 pKa = 3.42IPYY260 pKa = 10.13RR261 pKa = 11.84CDD263 pKa = 3.31PDD265 pKa = 3.71LVRR268 pKa = 11.84EE269 pKa = 4.05KK270 pKa = 10.05TINVDD275 pKa = 3.22PEE277 pKa = 4.29TIRR280 pKa = 11.84PHH282 pKa = 5.72IRR284 pKa = 11.84AILEE288 pKa = 4.01RR289 pKa = 11.84EE290 pKa = 4.01LVNRR294 pKa = 11.84NDD296 pKa = 3.94LPDD299 pKa = 4.47MEE301 pKa = 5.82DD302 pKa = 2.98FWTSRR307 pKa = 11.84WLWCVNGSQTSDD319 pKa = 2.83SDD321 pKa = 3.61KK322 pKa = 11.6ALGLDD327 pKa = 3.92LKK329 pKa = 9.88TLQTHH334 pKa = 5.07RR335 pKa = 11.84RR336 pKa = 11.84RR337 pKa = 11.84YY338 pKa = 9.75RR339 pKa = 11.84RR340 pKa = 11.84MAAEE344 pKa = 4.16ALVDD348 pKa = 4.0NPIPDD353 pKa = 3.79WDD355 pKa = 3.53GTTYY359 pKa = 10.89VSASSKK365 pKa = 11.11LEE367 pKa = 3.85VGKK370 pKa = 8.55TRR372 pKa = 11.84AIFACDD378 pKa = 3.1TRR380 pKa = 11.84SYY382 pKa = 11.04FAFSYY387 pKa = 10.91LLNQVQKK394 pKa = 9.94DD395 pKa = 3.44WRR397 pKa = 11.84NEE399 pKa = 3.78RR400 pKa = 11.84VLLDD404 pKa = 4.04PGNGGTVAMGARR416 pKa = 11.84LRR418 pKa = 11.84NAQKK422 pKa = 10.89GGGFNLMLDD431 pKa = 3.58YY432 pKa = 11.47DD433 pKa = 4.75DD434 pKa = 6.31FNSQHH439 pKa = 5.4STEE442 pKa = 3.96TMKK445 pKa = 10.63IVFEE449 pKa = 4.43EE450 pKa = 4.25TCKK453 pKa = 10.39IYY455 pKa = 10.56NAPAWYY461 pKa = 8.68TDD463 pKa = 3.74KK464 pKa = 11.22LCSSFDD470 pKa = 3.18KK471 pKa = 10.65MYY473 pKa = 10.49IRR475 pKa = 11.84QKK477 pKa = 10.92DD478 pKa = 3.84GLRR481 pKa = 11.84HH482 pKa = 4.58VAGTLMSGHH491 pKa = 7.15RR492 pKa = 11.84GTTFINSVLNAAYY505 pKa = 9.78LRR507 pKa = 11.84CGVGSGWFDD516 pKa = 3.12HH517 pKa = 6.82ALSLHH522 pKa = 6.26TGDD525 pKa = 5.04DD526 pKa = 3.7VYY528 pKa = 11.08IRR530 pKa = 11.84SNSRR534 pKa = 11.84SEE536 pKa = 3.82VSNILTRR543 pKa = 11.84AAEE546 pKa = 4.58FGCRR550 pKa = 11.84MNPTKK555 pKa = 10.6QSIGVKK561 pKa = 8.6NCEE564 pKa = 4.22FLRR567 pKa = 11.84CAYY570 pKa = 10.18NPYY573 pKa = 9.91YY574 pKa = 10.63AVGYY578 pKa = 9.26LCRR581 pKa = 11.84TVGTLVNGNWSGDD594 pKa = 3.66TPLTPHH600 pKa = 6.86EE601 pKa = 4.92ALTSLLTSLRR611 pKa = 11.84SLYY614 pKa = 10.3NRR616 pKa = 11.84SLGGGLGRR624 pKa = 11.84YY625 pKa = 7.71LASAIRR631 pKa = 11.84FRR633 pKa = 11.84TDD635 pKa = 3.71GISRR639 pKa = 11.84KK640 pKa = 9.28NLIGLLDD647 pKa = 3.55GRR649 pKa = 11.84IAIEE653 pKa = 4.82GGPCFNADD661 pKa = 2.99AKK663 pKa = 10.83IRR665 pKa = 11.84TCALKK670 pKa = 9.19NTIKK674 pKa = 10.63PSPLVDD680 pKa = 4.2KK681 pKa = 11.19NWDD684 pKa = 3.14AYY686 pKa = 9.38ATRR689 pKa = 11.84DD690 pKa = 3.65YY691 pKa = 10.82LVCHH695 pKa = 5.61TTDD698 pKa = 3.21LEE700 pKa = 4.55YY701 pKa = 11.12YY702 pKa = 10.25ALQNCGVSPTPLMVTTSYY720 pKa = 11.41EE721 pKa = 3.68KK722 pKa = 10.71GLNRR726 pKa = 11.84EE727 pKa = 4.14SEE729 pKa = 4.28EE730 pKa = 4.0VKK732 pKa = 10.42PLKK735 pKa = 9.77FCNMRR740 pKa = 11.84VSTARR745 pKa = 11.84GFATVSEE752 pKa = 4.18LLKK755 pKa = 10.59VDD757 pKa = 3.6TEE759 pKa = 4.71GGVLAHH765 pKa = 6.48YY766 pKa = 8.56PLLMLIKK773 pKa = 10.5DD774 pKa = 3.97SLSPSLIRR782 pKa = 11.84EE783 pKa = 4.07LCEE786 pKa = 3.84ADD788 pKa = 4.67GYY790 pKa = 9.29TPPSEE795 pKa = 4.53NIRR798 pKa = 11.84AYY800 pKa = 10.65AFGEE804 pKa = 4.01DD805 pKa = 3.86FRR807 pKa = 11.84THH809 pKa = 6.64NIIGRR814 pKa = 11.84LPYY817 pKa = 10.3SDD819 pKa = 4.93AAALQGKK826 pKa = 6.27TSCDD830 pKa = 3.59NIVADD835 pKa = 3.82YY836 pKa = 10.71KK837 pKa = 10.88IYY839 pKa = 10.97LL840 pKa = 3.86
Molecular weight: 93.79 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1583 |
743 |
840 |
791.5 |
85.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.36 ± 1.48 | 1.832 ± 0.518 |
5.433 ± 0.588 | 4.738 ± 0.166 |
3.285 ± 0.055 | 8.718 ± 1.314 |
2.653 ± 0.211 | 3.538 ± 0.58 |
2.59 ± 1.315 | 10.36 ± 0.921 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.021 ± 0.002 | 4.043 ± 0.558 |
6.001 ± 1.238 | 2.716 ± 0.017 |
7.138 ± 0.281 | 6.57 ± 0.017 |
6.57 ± 0.202 | 7.202 ± 0.969 |
1.263 ± 0.22 | 2.969 ± 0.652 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
