
Halosimplex carlsbadense 2-9-1
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Haloarculaceae; Halosimplex; Halosimplex carlsbadense
Average proteome isoelectric point is 4.83
Get precalculated fractions of proteins
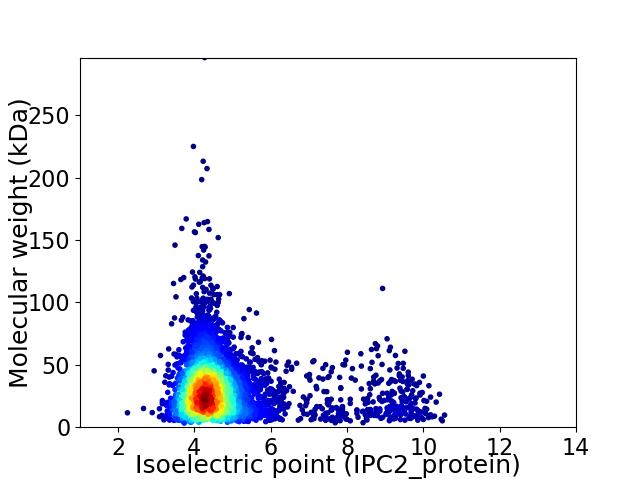
Virtual 2D-PAGE plot for 4458 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|M0CJ66|M0CJ66_9EURY Cupin 2 barrel domain-containing protein OS=Halosimplex carlsbadense 2-9-1 OX=797114 GN=C475_16216 PE=4 SV=1
MM1 pKa = 7.23ATDD4 pKa = 3.56ITRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84AIKK12 pKa = 10.55ALGLAGATGLAGCSSGGDD30 pKa = 3.53STATSGDD37 pKa = 3.7GGSGGSDD44 pKa = 3.01TVRR47 pKa = 11.84AAFVYY52 pKa = 10.15NSEE55 pKa = 4.47VGDD58 pKa = 4.03QGWSWAHH65 pKa = 5.41DD66 pKa = 3.46QGRR69 pKa = 11.84QAVAEE74 pKa = 4.1QYY76 pKa = 10.9DD77 pKa = 3.9WLEE80 pKa = 3.84TGYY83 pKa = 9.11TEE85 pKa = 5.15AVAPADD91 pKa = 3.43SRR93 pKa = 11.84TVFEE97 pKa = 5.41QYY99 pKa = 10.9VQDD102 pKa = 4.85DD103 pKa = 3.86YY104 pKa = 11.95DD105 pKa = 3.59IVFGNTFGYY114 pKa = 8.89MDD116 pKa = 3.67TMVSVAEE123 pKa = 4.34EE124 pKa = 4.27SPDD127 pKa = 3.4TLFEE131 pKa = 4.49HH132 pKa = 7.12CSGYY136 pKa = 8.06QTSEE140 pKa = 3.38NLGRR144 pKa = 11.84YY145 pKa = 8.5FGRR148 pKa = 11.84MYY150 pKa = 9.85QARR153 pKa = 11.84YY154 pKa = 8.48LAGIAAGMLTEE165 pKa = 5.0ADD167 pKa = 3.53SLGYY171 pKa = 10.01VAALPIAEE179 pKa = 4.64VIRR182 pKa = 11.84GINAFTLGARR192 pKa = 11.84SVNPDD197 pKa = 2.49ATTEE201 pKa = 4.02VRR203 pKa = 11.84WTNTWYY209 pKa = 10.16DD210 pKa = 3.58PPTEE214 pKa = 3.98QEE216 pKa = 4.03AANALIDD223 pKa = 3.59QGVDD227 pKa = 3.2VMAQHH232 pKa = 6.47QDD234 pKa = 2.71SAAAVQAAADD244 pKa = 3.73ADD246 pKa = 3.66IWATGYY252 pKa = 8.9DD253 pKa = 3.77APMLEE258 pKa = 4.25QGGDD262 pKa = 3.44NYY264 pKa = 9.6VTSPIWHH271 pKa = 6.41WEE273 pKa = 3.79EE274 pKa = 4.54FYY276 pKa = 11.34GPTLEE281 pKa = 4.38AVRR284 pKa = 11.84DD285 pKa = 4.2GSWEE289 pKa = 3.82SDD291 pKa = 3.75FYY293 pKa = 11.73FEE295 pKa = 5.24GLEE298 pKa = 3.89SDD300 pKa = 4.56IIGLSEE306 pKa = 4.41WGPEE310 pKa = 3.97VPDD313 pKa = 3.89DD314 pKa = 3.85VKK316 pKa = 11.5SSVEE320 pKa = 4.08EE321 pKa = 3.64THH323 pKa = 6.84SAIEE327 pKa = 4.5SGDD330 pKa = 3.47ADD332 pKa = 3.31VWADD336 pKa = 4.13SKK338 pKa = 11.36FADD341 pKa = 4.78ADD343 pKa = 3.97DD344 pKa = 3.97ATLFQDD350 pKa = 2.83MGSYY354 pKa = 10.31VEE356 pKa = 4.41GVEE359 pKa = 4.47GSVPEE364 pKa = 4.09
MM1 pKa = 7.23ATDD4 pKa = 3.56ITRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84AIKK12 pKa = 10.55ALGLAGATGLAGCSSGGDD30 pKa = 3.53STATSGDD37 pKa = 3.7GGSGGSDD44 pKa = 3.01TVRR47 pKa = 11.84AAFVYY52 pKa = 10.15NSEE55 pKa = 4.47VGDD58 pKa = 4.03QGWSWAHH65 pKa = 5.41DD66 pKa = 3.46QGRR69 pKa = 11.84QAVAEE74 pKa = 4.1QYY76 pKa = 10.9DD77 pKa = 3.9WLEE80 pKa = 3.84TGYY83 pKa = 9.11TEE85 pKa = 5.15AVAPADD91 pKa = 3.43SRR93 pKa = 11.84TVFEE97 pKa = 5.41QYY99 pKa = 10.9VQDD102 pKa = 4.85DD103 pKa = 3.86YY104 pKa = 11.95DD105 pKa = 3.59IVFGNTFGYY114 pKa = 8.89MDD116 pKa = 3.67TMVSVAEE123 pKa = 4.34EE124 pKa = 4.27SPDD127 pKa = 3.4TLFEE131 pKa = 4.49HH132 pKa = 7.12CSGYY136 pKa = 8.06QTSEE140 pKa = 3.38NLGRR144 pKa = 11.84YY145 pKa = 8.5FGRR148 pKa = 11.84MYY150 pKa = 9.85QARR153 pKa = 11.84YY154 pKa = 8.48LAGIAAGMLTEE165 pKa = 5.0ADD167 pKa = 3.53SLGYY171 pKa = 10.01VAALPIAEE179 pKa = 4.64VIRR182 pKa = 11.84GINAFTLGARR192 pKa = 11.84SVNPDD197 pKa = 2.49ATTEE201 pKa = 4.02VRR203 pKa = 11.84WTNTWYY209 pKa = 10.16DD210 pKa = 3.58PPTEE214 pKa = 3.98QEE216 pKa = 4.03AANALIDD223 pKa = 3.59QGVDD227 pKa = 3.2VMAQHH232 pKa = 6.47QDD234 pKa = 2.71SAAAVQAAADD244 pKa = 3.73ADD246 pKa = 3.66IWATGYY252 pKa = 8.9DD253 pKa = 3.77APMLEE258 pKa = 4.25QGGDD262 pKa = 3.44NYY264 pKa = 9.6VTSPIWHH271 pKa = 6.41WEE273 pKa = 3.79EE274 pKa = 4.54FYY276 pKa = 11.34GPTLEE281 pKa = 4.38AVRR284 pKa = 11.84DD285 pKa = 4.2GSWEE289 pKa = 3.82SDD291 pKa = 3.75FYY293 pKa = 11.73FEE295 pKa = 5.24GLEE298 pKa = 3.89SDD300 pKa = 4.56IIGLSEE306 pKa = 4.41WGPEE310 pKa = 3.97VPDD313 pKa = 3.89DD314 pKa = 3.85VKK316 pKa = 11.5SSVEE320 pKa = 4.08EE321 pKa = 3.64THH323 pKa = 6.84SAIEE327 pKa = 4.5SGDD330 pKa = 3.47ADD332 pKa = 3.31VWADD336 pKa = 4.13SKK338 pKa = 11.36FADD341 pKa = 4.78ADD343 pKa = 3.97DD344 pKa = 3.97ATLFQDD350 pKa = 2.83MGSYY354 pKa = 10.31VEE356 pKa = 4.41GVEE359 pKa = 4.47GSVPEE364 pKa = 4.09
Molecular weight: 39.18 kDa
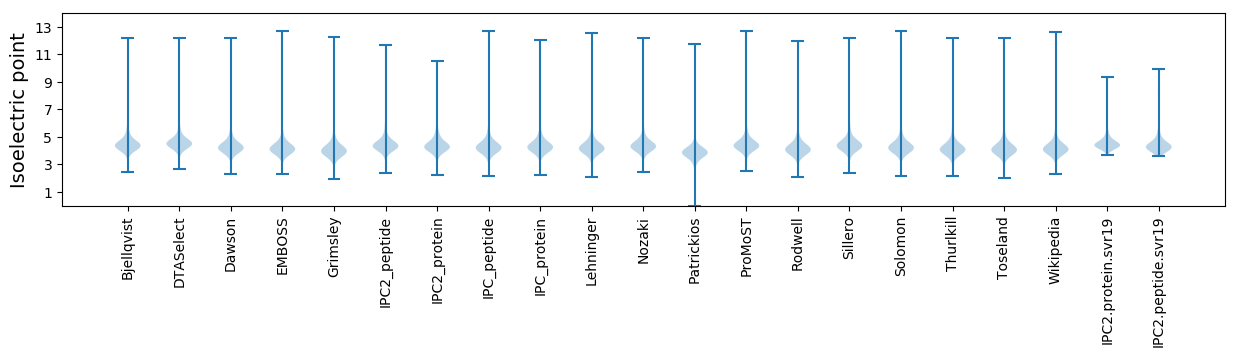
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|M0D1Y2|M0D1Y2_9EURY Uridine phosphorylase OS=Halosimplex carlsbadense 2-9-1 OX=797114 GN=C475_05145 PE=3 SV=1
MM1 pKa = 6.85YY2 pKa = 10.42AEE4 pKa = 4.82PARR7 pKa = 11.84KK8 pKa = 6.08TTAPSAVNSKK18 pKa = 10.14KK19 pKa = 10.36PNARR23 pKa = 11.84RR24 pKa = 11.84PGYY27 pKa = 10.39SPARR31 pKa = 11.84LAAVALARR39 pKa = 11.84RR40 pKa = 11.84FVEE43 pKa = 4.29VPMSVSIPPICEE55 pKa = 4.15AYY57 pKa = 10.38DD58 pKa = 3.51SGISSFDD65 pKa = 3.48GEE67 pKa = 4.8VFPSLATSVTTGTSTATTGVLLTNPEE93 pKa = 4.26TGPATPTVASSCRR106 pKa = 11.84SRR108 pKa = 11.84LSPANAAIRR117 pKa = 11.84RR118 pKa = 11.84PRR120 pKa = 11.84IWTSPVRR127 pKa = 11.84LIPALKK133 pKa = 9.92ISIASTVIVAGLEE146 pKa = 4.04NPEE149 pKa = 4.08IPSLGEE155 pKa = 3.69IPVHH159 pKa = 5.62GRR161 pKa = 11.84RR162 pKa = 11.84TTSVTMIAIAVTSIGSHH179 pKa = 5.18SVTNSAMAPRR189 pKa = 11.84TTSPTTTMSTVTPAGADD206 pKa = 3.4GCRR209 pKa = 11.84SVTEE213 pKa = 4.2TEE215 pKa = 3.83RR216 pKa = 11.84LTRR219 pKa = 3.93
MM1 pKa = 6.85YY2 pKa = 10.42AEE4 pKa = 4.82PARR7 pKa = 11.84KK8 pKa = 6.08TTAPSAVNSKK18 pKa = 10.14KK19 pKa = 10.36PNARR23 pKa = 11.84RR24 pKa = 11.84PGYY27 pKa = 10.39SPARR31 pKa = 11.84LAAVALARR39 pKa = 11.84RR40 pKa = 11.84FVEE43 pKa = 4.29VPMSVSIPPICEE55 pKa = 4.15AYY57 pKa = 10.38DD58 pKa = 3.51SGISSFDD65 pKa = 3.48GEE67 pKa = 4.8VFPSLATSVTTGTSTATTGVLLTNPEE93 pKa = 4.26TGPATPTVASSCRR106 pKa = 11.84SRR108 pKa = 11.84LSPANAAIRR117 pKa = 11.84RR118 pKa = 11.84PRR120 pKa = 11.84IWTSPVRR127 pKa = 11.84LIPALKK133 pKa = 9.92ISIASTVIVAGLEE146 pKa = 4.04NPEE149 pKa = 4.08IPSLGEE155 pKa = 3.69IPVHH159 pKa = 5.62GRR161 pKa = 11.84RR162 pKa = 11.84TTSVTMIAIAVTSIGSHH179 pKa = 5.18SVTNSAMAPRR189 pKa = 11.84TTSPTTTMSTVTPAGADD206 pKa = 3.4GCRR209 pKa = 11.84SVTEE213 pKa = 4.2TEE215 pKa = 3.83RR216 pKa = 11.84LTRR219 pKa = 3.93
Molecular weight: 22.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1310473 |
29 |
2791 |
294.0 |
31.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.636 ± 0.064 | 0.683 ± 0.012 |
8.912 ± 0.045 | 8.319 ± 0.055 |
3.222 ± 0.025 | 9.066 ± 0.042 |
1.887 ± 0.019 | 3.371 ± 0.03 |
1.496 ± 0.022 | 8.564 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.646 ± 0.018 | 2.15 ± 0.026 |
4.79 ± 0.027 | 2.304 ± 0.02 |
6.847 ± 0.042 | 5.448 ± 0.036 |
6.359 ± 0.036 | 9.418 ± 0.037 |
1.221 ± 0.016 | 2.656 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
