
Lachnospiraceae bacterium KH1T2
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; unclassified Lachnospiraceae
Average proteome isoelectric point is 5.91
Get precalculated fractions of proteins
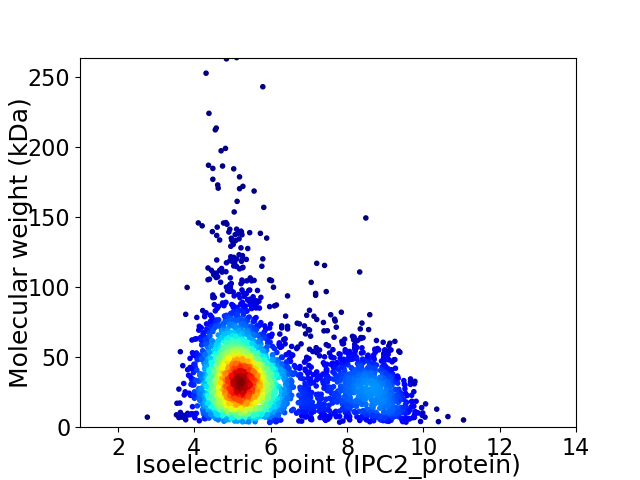
Virtual 2D-PAGE plot for 3398 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I4E4M0|A0A1I4E4M0_9FIRM WG containing repeat-containing protein OS=Lachnospiraceae bacterium KH1T2 OX=1855374 GN=SAMN05216390_10756 PE=4 SV=1
MM1 pKa = 7.52LKK3 pKa = 10.58KK4 pKa = 10.51NLIKK8 pKa = 10.76KK9 pKa = 10.17LGISAIAVFMAAMLITGCGNTVTDD33 pKa = 4.32SAPTTATSSTSVAEE47 pKa = 4.18ADD49 pKa = 3.75PSGEE53 pKa = 3.99VDD55 pKa = 3.8KK56 pKa = 11.57DD57 pKa = 3.78MPDD60 pKa = 3.21GQAPDD65 pKa = 3.86GQGGPGGQAPDD76 pKa = 3.79GQGGPGGQSSAPTEE90 pKa = 3.67YY91 pKa = 10.93AAANEE96 pKa = 4.24YY97 pKa = 10.3TSDD100 pKa = 3.64EE101 pKa = 4.44TVDD104 pKa = 3.77DD105 pKa = 4.23LTVEE109 pKa = 4.36STGSDD114 pKa = 3.14EE115 pKa = 4.7NAVHH119 pKa = 6.16VLNGSQVIFNNLMISRR135 pKa = 11.84NSSDD139 pKa = 3.14SSGGDD144 pKa = 2.95SASFYY149 pKa = 11.36GVGASLLTTDD159 pKa = 3.74GTAIVNGGTITSDD172 pKa = 3.27SKK174 pKa = 11.53GGAGLFAYY182 pKa = 10.28GDD184 pKa = 3.78GTIYY188 pKa = 10.42AQNVKK193 pKa = 9.73IDD195 pKa = 3.9TKK197 pKa = 10.78QDD199 pKa = 3.08TSGGVHH205 pKa = 6.49AAGGGTFYY213 pKa = 10.99GWDD216 pKa = 3.23LTVNTEE222 pKa = 4.38GEE224 pKa = 3.97SSAAIRR230 pKa = 11.84SDD232 pKa = 3.19RR233 pKa = 11.84GGGTMVIDD241 pKa = 4.31GGTYY245 pKa = 7.55TANGSGSPALYY256 pKa = 8.59CTADD260 pKa = 3.03ISVNNATLEE269 pKa = 4.23ATGSEE274 pKa = 4.9GVCLEE279 pKa = 4.04GLNTTRR285 pKa = 11.84LFDD288 pKa = 3.82TDD290 pKa = 4.57LTCDD294 pKa = 4.17MPDD297 pKa = 3.18QEE299 pKa = 5.02QNNNLSWSVIVYY311 pKa = 10.21QSMSGDD317 pKa = 3.83SEE319 pKa = 4.47VGAGTFDD326 pKa = 3.47MVGGSITSKK335 pKa = 10.82NGGLFYY341 pKa = 8.78TTNTEE346 pKa = 4.3SNFYY350 pKa = 10.67LSGVNITTADD360 pKa = 3.39DD361 pKa = 3.92CEE363 pKa = 4.44FFLRR367 pKa = 11.84ATGNANGRR375 pKa = 11.84GWGTTGEE382 pKa = 4.28NGADD386 pKa = 3.72CNFTADD392 pKa = 3.93DD393 pKa = 3.75QDD395 pKa = 3.56MKK397 pKa = 11.65GDD399 pKa = 3.91VQWDD403 pKa = 4.17SISKK407 pKa = 10.47LDD409 pKa = 3.86FYY411 pKa = 10.65MLNGSTLQGAFVDD424 pKa = 4.69DD425 pKa = 4.34EE426 pKa = 4.82SEE428 pKa = 4.11AGNGSDD434 pKa = 3.4EE435 pKa = 4.82GYY437 pKa = 11.2ANLYY441 pKa = 9.52IDD443 pKa = 5.5SSSTWTVTGDD453 pKa = 3.34STLTALHH460 pKa = 6.19NAGKK464 pKa = 10.14IVDD467 pKa = 3.76EE468 pKa = 4.81SGNSVTVKK476 pKa = 9.73GTDD479 pKa = 3.03GTVYY483 pKa = 10.8VEE485 pKa = 4.53GDD487 pKa = 3.42SEE489 pKa = 4.36YY490 pKa = 10.68TITVDD495 pKa = 5.11SYY497 pKa = 11.15DD498 pKa = 3.82TEE500 pKa = 6.2ADD502 pKa = 3.18TSEE505 pKa = 4.92ALTAPVWSDD514 pKa = 3.32YY515 pKa = 11.37AVEE518 pKa = 4.44KK519 pKa = 10.98PEE521 pKa = 4.21ALQQ524 pKa = 3.81
MM1 pKa = 7.52LKK3 pKa = 10.58KK4 pKa = 10.51NLIKK8 pKa = 10.76KK9 pKa = 10.17LGISAIAVFMAAMLITGCGNTVTDD33 pKa = 4.32SAPTTATSSTSVAEE47 pKa = 4.18ADD49 pKa = 3.75PSGEE53 pKa = 3.99VDD55 pKa = 3.8KK56 pKa = 11.57DD57 pKa = 3.78MPDD60 pKa = 3.21GQAPDD65 pKa = 3.86GQGGPGGQAPDD76 pKa = 3.79GQGGPGGQSSAPTEE90 pKa = 3.67YY91 pKa = 10.93AAANEE96 pKa = 4.24YY97 pKa = 10.3TSDD100 pKa = 3.64EE101 pKa = 4.44TVDD104 pKa = 3.77DD105 pKa = 4.23LTVEE109 pKa = 4.36STGSDD114 pKa = 3.14EE115 pKa = 4.7NAVHH119 pKa = 6.16VLNGSQVIFNNLMISRR135 pKa = 11.84NSSDD139 pKa = 3.14SSGGDD144 pKa = 2.95SASFYY149 pKa = 11.36GVGASLLTTDD159 pKa = 3.74GTAIVNGGTITSDD172 pKa = 3.27SKK174 pKa = 11.53GGAGLFAYY182 pKa = 10.28GDD184 pKa = 3.78GTIYY188 pKa = 10.42AQNVKK193 pKa = 9.73IDD195 pKa = 3.9TKK197 pKa = 10.78QDD199 pKa = 3.08TSGGVHH205 pKa = 6.49AAGGGTFYY213 pKa = 10.99GWDD216 pKa = 3.23LTVNTEE222 pKa = 4.38GEE224 pKa = 3.97SSAAIRR230 pKa = 11.84SDD232 pKa = 3.19RR233 pKa = 11.84GGGTMVIDD241 pKa = 4.31GGTYY245 pKa = 7.55TANGSGSPALYY256 pKa = 8.59CTADD260 pKa = 3.03ISVNNATLEE269 pKa = 4.23ATGSEE274 pKa = 4.9GVCLEE279 pKa = 4.04GLNTTRR285 pKa = 11.84LFDD288 pKa = 3.82TDD290 pKa = 4.57LTCDD294 pKa = 4.17MPDD297 pKa = 3.18QEE299 pKa = 5.02QNNNLSWSVIVYY311 pKa = 10.21QSMSGDD317 pKa = 3.83SEE319 pKa = 4.47VGAGTFDD326 pKa = 3.47MVGGSITSKK335 pKa = 10.82NGGLFYY341 pKa = 8.78TTNTEE346 pKa = 4.3SNFYY350 pKa = 10.67LSGVNITTADD360 pKa = 3.39DD361 pKa = 3.92CEE363 pKa = 4.44FFLRR367 pKa = 11.84ATGNANGRR375 pKa = 11.84GWGTTGEE382 pKa = 4.28NGADD386 pKa = 3.72CNFTADD392 pKa = 3.93DD393 pKa = 3.75QDD395 pKa = 3.56MKK397 pKa = 11.65GDD399 pKa = 3.91VQWDD403 pKa = 4.17SISKK407 pKa = 10.47LDD409 pKa = 3.86FYY411 pKa = 10.65MLNGSTLQGAFVDD424 pKa = 4.69DD425 pKa = 4.34EE426 pKa = 4.82SEE428 pKa = 4.11AGNGSDD434 pKa = 3.4EE435 pKa = 4.82GYY437 pKa = 11.2ANLYY441 pKa = 9.52IDD443 pKa = 5.5SSSTWTVTGDD453 pKa = 3.34STLTALHH460 pKa = 6.19NAGKK464 pKa = 10.14IVDD467 pKa = 3.76EE468 pKa = 4.81SGNSVTVKK476 pKa = 9.73GTDD479 pKa = 3.03GTVYY483 pKa = 10.8VEE485 pKa = 4.53GDD487 pKa = 3.42SEE489 pKa = 4.36YY490 pKa = 10.68TITVDD495 pKa = 5.11SYY497 pKa = 11.15DD498 pKa = 3.82TEE500 pKa = 6.2ADD502 pKa = 3.18TSEE505 pKa = 4.92ALTAPVWSDD514 pKa = 3.32YY515 pKa = 11.37AVEE518 pKa = 4.44KK519 pKa = 10.98PEE521 pKa = 4.21ALQQ524 pKa = 3.81
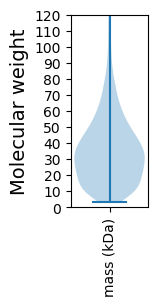
Molecular weight: 53.85 kDa
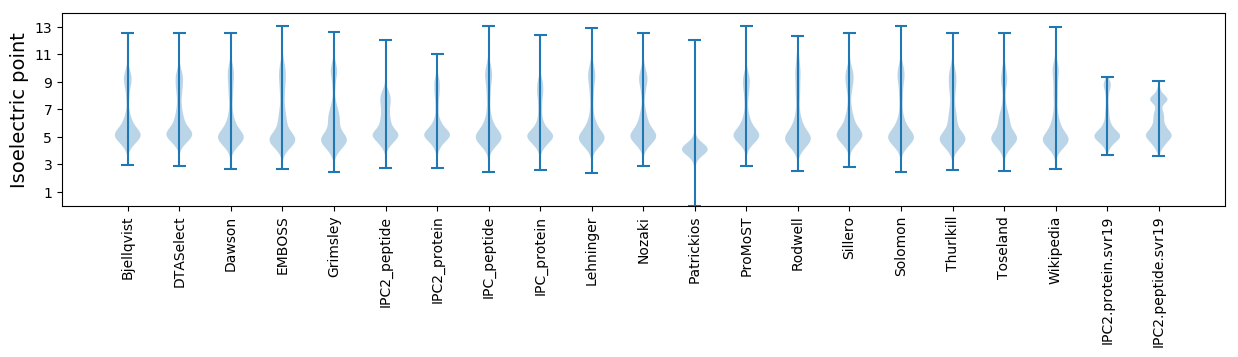
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I4G327|A0A1I4G327_9FIRM Uncharacterized protein OS=Lachnospiraceae bacterium KH1T2 OX=1855374 GN=SAMN05216390_11566 PE=4 SV=1
MM1 pKa = 7.45WMTYY5 pKa = 7.14QPKK8 pKa = 9.66KK9 pKa = 8.41RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.87GFRR19 pKa = 11.84ARR21 pKa = 11.84MATPGGRR28 pKa = 11.84KK29 pKa = 8.94VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.66GRR39 pKa = 11.84AKK41 pKa = 10.06LTVV44 pKa = 3.04
MM1 pKa = 7.45WMTYY5 pKa = 7.14QPKK8 pKa = 9.66KK9 pKa = 8.41RR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 9.17VHH16 pKa = 5.87GFRR19 pKa = 11.84ARR21 pKa = 11.84MATPGGRR28 pKa = 11.84KK29 pKa = 8.94VLAARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.66GRR39 pKa = 11.84AKK41 pKa = 10.06LTVV44 pKa = 3.04
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1154851 |
30 |
2372 |
339.9 |
38.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.431 ± 0.047 | 1.36 ± 0.016 |
6.33 ± 0.038 | 7.698 ± 0.051 |
4.308 ± 0.031 | 6.761 ± 0.039 |
1.629 ± 0.018 | 7.814 ± 0.041 |
7.436 ± 0.036 | 8.258 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.974 ± 0.019 | 4.983 ± 0.031 |
2.967 ± 0.022 | 2.482 ± 0.023 |
4.064 ± 0.033 | 6.395 ± 0.037 |
5.177 ± 0.033 | 6.74 ± 0.033 |
0.873 ± 0.015 | 4.319 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
