
Chimaeribacter arupi
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Yersiniaceae; Chimaeribacter
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
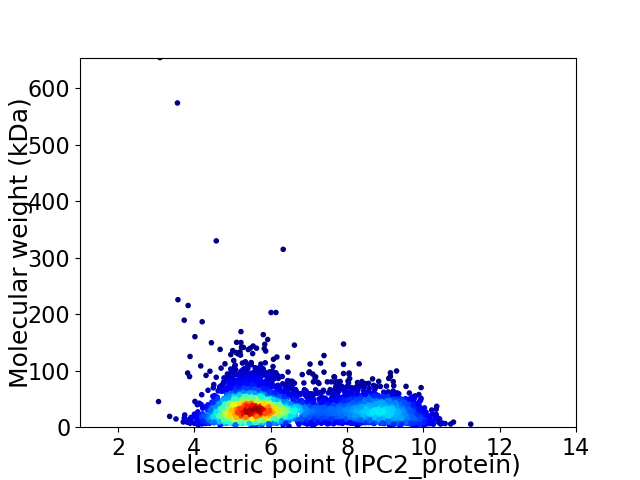
Virtual 2D-PAGE plot for 4159 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2N5EPZ0|A0A2N5EPZ0_9GAMM EamA family transporter OS=Chimaeribacter arupi OX=2060066 GN=CYR34_07315 PE=4 SV=1
MM1 pKa = 7.38NNGDD5 pKa = 3.62NGMDD9 pKa = 3.97IIYY12 pKa = 10.7SLLNKK17 pKa = 8.54RR18 pKa = 11.84QSMTTATLFSRR29 pKa = 11.84QTGNLISQIIDD40 pKa = 3.22ASQVISLTEE49 pKa = 3.77PSVITINNARR59 pKa = 11.84GWATGYY65 pKa = 10.4SRR67 pKa = 11.84SGDD70 pKa = 3.88DD71 pKa = 5.43LSLATQDD78 pKa = 3.53GTTLQYY84 pKa = 11.2RR85 pKa = 11.84HH86 pKa = 6.16FFAAGSAGTLVFDD99 pKa = 5.5DD100 pKa = 4.75GSGPATQAVFSHH112 pKa = 7.23DD113 pKa = 3.97AGSSSLTPVFKK124 pKa = 10.42AYY126 pKa = 9.52HH127 pKa = 6.23APVEE131 pKa = 4.2AAGAARR137 pKa = 11.84STQASAADD145 pKa = 3.53INAVADD151 pKa = 3.47VDD153 pKa = 4.58AADD156 pKa = 4.35TPTITINTFAGDD168 pKa = 3.75NIINYY173 pKa = 10.21AEE175 pKa = 4.27GQVEE179 pKa = 4.29QVLSGTTTGIEE190 pKa = 4.07AGQSLTVAFNSIVSFWTEE208 pKa = 3.71VQADD212 pKa = 4.35GSWQLTLNAASMYY225 pKa = 10.93DD226 pKa = 3.37LLRR229 pKa = 11.84YY230 pKa = 9.9GSTEE234 pKa = 3.34ITAEE238 pKa = 4.05SANAAGEE245 pKa = 4.16EE246 pKa = 4.36ATVSVPFQVLSDD258 pKa = 3.83WPSVTLNAFAGDD270 pKa = 3.95NVISGPEE277 pKa = 3.83DD278 pKa = 3.13QATQILSGSTSRR290 pKa = 11.84VEE292 pKa = 4.09AGQTVTLTLNGNTYY306 pKa = 8.73TATVDD311 pKa = 3.05ADD313 pKa = 4.45GYY315 pKa = 10.27WRR317 pKa = 11.84TSVPAADD324 pKa = 3.81VAALPDD330 pKa = 4.03GAQLVATVTNALGRR344 pKa = 11.84EE345 pKa = 4.26AGDD348 pKa = 3.66SATLTVNSSLPSLTVDD364 pKa = 4.31PFATDD369 pKa = 3.22NVLSDD374 pKa = 5.16AEE376 pKa = 4.17AQSAQTLSGTTTGVEE391 pKa = 3.68AGQAVTVTLNEE402 pKa = 3.81ITYY405 pKa = 8.07STTVAADD412 pKa = 3.71GSFSLSVPSADD423 pKa = 5.09LLALYY428 pKa = 10.32SGQTTIAARR437 pKa = 11.84AANLAGQEE445 pKa = 4.37TQGSEE450 pKa = 3.55ILTVDD455 pKa = 4.77FATARR460 pKa = 11.84LVVDD464 pKa = 4.72PVAGDD469 pKa = 3.5NYY471 pKa = 10.3INASEE476 pKa = 4.05SSYY479 pKa = 11.35LVLSGTTVDD488 pKa = 3.83VPAGSWVVITFNDD501 pKa = 4.06RR502 pKa = 11.84IFRR505 pKa = 11.84AEE507 pKa = 3.96VEE509 pKa = 4.46ADD511 pKa = 4.04GSWSTYY517 pKa = 10.53VSDD520 pKa = 4.46LVVNSFPDD528 pKa = 3.57GQYY531 pKa = 10.46PITFSAPGSAPVTLDD546 pKa = 2.86VGLYY550 pKa = 9.84IDD552 pKa = 4.82NQAQLNVQIDD562 pKa = 4.02TLFGDD567 pKa = 4.79DD568 pKa = 3.8VLTPGEE574 pKa = 4.15TLQDD578 pKa = 3.49QVLTGTTGITGAGQTISAEE597 pKa = 4.26LEE599 pKa = 4.48GVTHH603 pKa = 6.89TGSVDD608 pKa = 3.2NDD610 pKa = 4.08GNWTITVPAAALQALQNPNGYY631 pKa = 10.36VYY633 pKa = 10.92VQAADD638 pKa = 3.58LAGNSIAVGRR648 pKa = 11.84SFEE651 pKa = 4.91LVFSPATLTIDD662 pKa = 5.46PITGDD667 pKa = 3.91NILTAAEE674 pKa = 4.2TATPLTVSGVSYY686 pKa = 10.23RR687 pKa = 11.84VPEE690 pKa = 4.32GVTITVTLNGKK701 pKa = 9.56EE702 pKa = 4.13YY703 pKa = 7.83TTTAPGNDD711 pKa = 3.24TWSVQVPAADD721 pKa = 3.97LAGLTDD727 pKa = 3.91GVYY730 pKa = 9.55TVTAATINIDD740 pKa = 3.91LEE742 pKa = 4.51PVTTQQDD749 pKa = 3.89LTVDD753 pKa = 4.55LAPAPAFSVDD763 pKa = 3.88PVTGDD768 pKa = 3.9DD769 pKa = 3.4ILYY772 pKa = 10.17VDD774 pKa = 4.11EE775 pKa = 4.55TSGEE779 pKa = 3.96IVITGRR785 pKa = 11.84APVSAVGGSVTVTAFDD801 pKa = 4.04DD802 pKa = 3.54PAYY805 pKa = 10.44RR806 pKa = 11.84YY807 pKa = 7.59TGTVAADD814 pKa = 3.79GTWQVTVPEE823 pKa = 4.39YY824 pKa = 11.36NFGGTSNGVYY834 pKa = 9.85TLVAEE839 pKa = 4.71LTAPDD844 pKa = 3.91GTTTTTTHH852 pKa = 7.66DD853 pKa = 2.94ITINVWPFSGDD864 pKa = 3.29FYY866 pKa = 11.49FDD868 pKa = 4.79DD869 pKa = 4.02FTGDD873 pKa = 3.23STLTAEE879 pKa = 5.45EE880 pKa = 4.22RR881 pKa = 11.84TQDD884 pKa = 3.45QVFSGTFASEE894 pKa = 4.09EE895 pKa = 4.09NDD897 pKa = 3.29VSGLGITVYY906 pKa = 9.7MHH908 pKa = 7.42DD909 pKa = 4.17NDD911 pKa = 4.37GQTFAYY917 pKa = 9.41TGTTDD922 pKa = 3.67GNGRR926 pKa = 11.84WSVTIPAADD935 pKa = 3.64LQQLSEE941 pKa = 4.02GWSYY945 pKa = 7.65MTATATDD952 pKa = 3.42AAGNGASSSRR962 pKa = 11.84EE963 pKa = 3.22ADD965 pKa = 3.78FAVLPATGGTITVNPIAGDD984 pKa = 3.62NVLTADD990 pKa = 4.93EE991 pKa = 4.81ISDD994 pKa = 3.69GLTISGSSTYY1004 pKa = 9.8MGEE1007 pKa = 4.17RR1008 pKa = 11.84YY1009 pKa = 7.47TQIEE1013 pKa = 4.17VEE1015 pKa = 4.15INGTLRR1021 pKa = 11.84IYY1023 pKa = 11.25ADD1025 pKa = 4.12YY1026 pKa = 10.95EE1027 pKa = 4.49SDD1029 pKa = 2.92WSVTFSRR1036 pKa = 11.84EE1037 pKa = 4.0FIATLPDD1044 pKa = 3.67GPLTVTASGYY1054 pKa = 10.15DD1055 pKa = 3.34LSGNRR1060 pKa = 11.84VTTTATLEE1068 pKa = 4.25VNLSDD1073 pKa = 4.91VSTLTLDD1080 pKa = 3.63PVTGDD1085 pKa = 3.92DD1086 pKa = 4.89VISADD1091 pKa = 3.57EE1092 pKa = 4.33AGGEE1096 pKa = 4.07IVITGRR1102 pKa = 11.84APLSTQGGTVTVTAFDD1118 pKa = 4.13DD1119 pKa = 3.53PAYY1122 pKa = 10.4RR1123 pKa = 11.84YY1124 pKa = 6.88TATVEE1129 pKa = 4.89EE1130 pKa = 5.09GGTWQATVPAGAFSGRR1146 pKa = 11.84ANGVYY1151 pKa = 9.79TLVAEE1156 pKa = 4.71LTAPDD1161 pKa = 3.88GTVTTLNHH1169 pKa = 6.92DD1170 pKa = 3.68VTSNQLPFSGGFAFYY1185 pKa = 11.02VFAGDD1190 pKa = 3.78DD1191 pKa = 3.45VLNAEE1196 pKa = 4.87EE1197 pKa = 4.0QAQDD1201 pKa = 3.46QILEE1205 pKa = 4.36GEE1207 pKa = 4.52VFSDD1211 pKa = 4.67DD1212 pKa = 3.79GGNVAGLAVTIQMTTFDD1229 pKa = 4.33GQQFTYY1235 pKa = 8.05TTTTGEE1241 pKa = 3.69NGYY1244 pKa = 8.39WQVTLPSSDD1253 pKa = 4.43LEE1255 pKa = 3.95QLRR1258 pKa = 11.84NGWVNLVGTVADD1270 pKa = 4.16GAGNTLSNEE1279 pKa = 4.15SQFTVLAGIGTLTINPIAGDD1299 pKa = 3.68NVLTADD1305 pKa = 4.31EE1306 pKa = 4.73IGNGLVVSGSGEE1318 pKa = 4.1FLDD1321 pKa = 4.23MPLSLEE1327 pKa = 3.99VTINGVSQGFSDD1339 pKa = 4.83YY1340 pKa = 11.38SSDD1343 pKa = 2.81WSVTFTPQQLAGFPDD1358 pKa = 4.56GPLTVTVTAYY1368 pKa = 10.36DD1369 pKa = 3.37IEE1371 pKa = 4.89FNLITATSTLTVDD1384 pKa = 5.3LGDD1387 pKa = 4.16LQPLTFNPFGGDD1399 pKa = 3.55DD1400 pKa = 3.7NYY1402 pKa = 11.42LDD1404 pKa = 4.19EE1405 pKa = 6.73DD1406 pKa = 4.18EE1407 pKa = 5.81LEE1409 pKa = 4.27SDD1411 pKa = 4.1QLLSGQAGNLAAGSIVTLTLGEE1433 pKa = 4.01FTYY1436 pKa = 10.56QAEE1439 pKa = 4.42VQADD1443 pKa = 3.89HH1444 pKa = 6.48SWSVLIPSADD1454 pKa = 3.86LQTLPNDD1461 pKa = 3.22ATTGISATVTDD1472 pKa = 4.25ADD1474 pKa = 4.36GNLLAAGSHH1483 pKa = 5.26TFLLYY1488 pKa = 8.6RR1489 pKa = 11.84TPGEE1493 pKa = 4.1LNIDD1497 pKa = 3.73TLAGNGILTADD1508 pKa = 4.12EE1509 pKa = 4.25ATVPLVISGTITNGRR1524 pKa = 11.84VGAAVTVDD1532 pKa = 4.07LNGHH1536 pKa = 7.27LYY1538 pKa = 8.17QTTVTASDD1546 pKa = 4.28GSWQVSVPVEE1556 pKa = 4.34DD1557 pKa = 4.74LAQLGNTEE1565 pKa = 4.25YY1566 pKa = 10.01WVSATSTDD1574 pKa = 3.83LSGNEE1579 pKa = 3.75INGLNLLVVDD1589 pKa = 5.45LYY1591 pKa = 11.84VPVITLDD1598 pKa = 4.31PIAGDD1603 pKa = 3.86NIINAAEE1610 pKa = 3.92AAAGVVLSGQSSEE1623 pKa = 4.17AGGVVSVQLNGVAYY1637 pKa = 9.05EE1638 pKa = 4.16AGVDD1642 pKa = 4.83EE1643 pKa = 4.73EE1644 pKa = 5.53GSWQLALPNSALAEE1658 pKa = 4.33LEE1660 pKa = 4.07DD1661 pKa = 3.94GRR1663 pKa = 11.84YY1664 pKa = 7.65TLTLSQTGFNGQTTTVTEE1682 pKa = 4.2SLLLDD1687 pKa = 4.37ADD1689 pKa = 3.96PANRR1693 pKa = 11.84PVLRR1697 pKa = 11.84IHH1699 pKa = 7.15TISHH1703 pKa = 6.77DD1704 pKa = 3.65NVLNGAEE1711 pKa = 4.21LQSDD1715 pKa = 3.85QIITGSSEE1723 pKa = 3.94NVEE1726 pKa = 4.29SGQTLSLKK1734 pKa = 10.42IGGVTYY1740 pKa = 7.72TTQVQPGGGWSLIVPAEE1757 pKa = 4.11DD1758 pKa = 3.93LAALGDD1764 pKa = 3.86GGLTLRR1770 pKa = 11.84AGVTDD1775 pKa = 3.93ASGNRR1780 pKa = 11.84ATASRR1785 pKa = 11.84NLTIKK1790 pKa = 10.26SDD1792 pKa = 3.85RR1793 pKa = 11.84DD1794 pKa = 3.58GLSIDD1799 pKa = 4.35PVTGDD1804 pKa = 3.77NLINAADD1811 pKa = 3.8AEE1813 pKa = 4.55GAITLTGHH1821 pKa = 6.04TDD1823 pKa = 3.71GVRR1826 pKa = 11.84HH1827 pKa = 5.99GATVRR1832 pKa = 11.84LTLNGEE1838 pKa = 4.03HH1839 pKa = 5.76YY1840 pKa = 7.81TAKK1843 pKa = 10.3VGKK1846 pKa = 9.86DD1847 pKa = 3.57GSWSAKK1853 pKa = 9.74VPAADD1858 pKa = 3.35VAALEE1863 pKa = 4.61EE1864 pKa = 5.01GSATLTACVKK1874 pKa = 9.85SACGQMLYY1882 pKa = 10.91AKK1884 pKa = 10.34AVLTVDD1890 pKa = 3.31TRR1892 pKa = 11.84APEE1895 pKa = 4.13LALLPVTGDD1904 pKa = 3.56NLISLQEE1911 pKa = 4.04AQAGFALTGSAGIAAAGLLVLVVLNGVEE1939 pKa = 4.19YY1940 pKa = 10.2QGKK1943 pKa = 8.51VQASGDD1949 pKa = 3.47WTVAIPAGTLADD1961 pKa = 4.17QPAGDD1966 pKa = 4.01YY1967 pKa = 10.28PLTVSLTDD1975 pKa = 3.26TAGNLTSLTSVVTLEE1990 pKa = 4.3AAPDD1994 pKa = 3.56ADD1996 pKa = 3.75ATEE1999 pKa = 4.41TAALTVTSQTVAASADD2015 pKa = 3.7STLTSALAVPDD2026 pKa = 4.19TATDD2030 pKa = 3.71TADD2033 pKa = 2.94GTYY2036 pKa = 10.45AIGGQTLTLTEE2047 pKa = 4.37SGGEE2051 pKa = 3.91ALGGSGNDD2059 pKa = 3.53TILLNTLDD2067 pKa = 4.37FLHH2070 pKa = 7.2IDD2072 pKa = 4.04GGSGTDD2078 pKa = 3.35TLLLAGSDD2086 pKa = 3.36QHH2088 pKa = 8.28LDD2090 pKa = 3.34LTTLGLKK2097 pKa = 9.77IEE2099 pKa = 4.97NIDD2102 pKa = 4.76IFDD2105 pKa = 4.87LGNGSNSLTLGLHH2118 pKa = 5.09EE2119 pKa = 5.32AEE2121 pKa = 4.65TVRR2124 pKa = 11.84DD2125 pKa = 3.94TPEE2128 pKa = 3.39EE2129 pKa = 4.02SLFIRR2134 pKa = 11.84GQGGSQLTLAGDD2146 pKa = 4.13NTWEE2150 pKa = 3.97TSGQRR2155 pKa = 11.84EE2156 pKa = 4.21VGGLLFDD2163 pKa = 4.44VYY2165 pKa = 10.61HH2166 pKa = 5.88ATGLEE2171 pKa = 4.21SADD2174 pKa = 4.35LLVQQGILVQQGG2186 pKa = 2.84
MM1 pKa = 7.38NNGDD5 pKa = 3.62NGMDD9 pKa = 3.97IIYY12 pKa = 10.7SLLNKK17 pKa = 8.54RR18 pKa = 11.84QSMTTATLFSRR29 pKa = 11.84QTGNLISQIIDD40 pKa = 3.22ASQVISLTEE49 pKa = 3.77PSVITINNARR59 pKa = 11.84GWATGYY65 pKa = 10.4SRR67 pKa = 11.84SGDD70 pKa = 3.88DD71 pKa = 5.43LSLATQDD78 pKa = 3.53GTTLQYY84 pKa = 11.2RR85 pKa = 11.84HH86 pKa = 6.16FFAAGSAGTLVFDD99 pKa = 5.5DD100 pKa = 4.75GSGPATQAVFSHH112 pKa = 7.23DD113 pKa = 3.97AGSSSLTPVFKK124 pKa = 10.42AYY126 pKa = 9.52HH127 pKa = 6.23APVEE131 pKa = 4.2AAGAARR137 pKa = 11.84STQASAADD145 pKa = 3.53INAVADD151 pKa = 3.47VDD153 pKa = 4.58AADD156 pKa = 4.35TPTITINTFAGDD168 pKa = 3.75NIINYY173 pKa = 10.21AEE175 pKa = 4.27GQVEE179 pKa = 4.29QVLSGTTTGIEE190 pKa = 4.07AGQSLTVAFNSIVSFWTEE208 pKa = 3.71VQADD212 pKa = 4.35GSWQLTLNAASMYY225 pKa = 10.93DD226 pKa = 3.37LLRR229 pKa = 11.84YY230 pKa = 9.9GSTEE234 pKa = 3.34ITAEE238 pKa = 4.05SANAAGEE245 pKa = 4.16EE246 pKa = 4.36ATVSVPFQVLSDD258 pKa = 3.83WPSVTLNAFAGDD270 pKa = 3.95NVISGPEE277 pKa = 3.83DD278 pKa = 3.13QATQILSGSTSRR290 pKa = 11.84VEE292 pKa = 4.09AGQTVTLTLNGNTYY306 pKa = 8.73TATVDD311 pKa = 3.05ADD313 pKa = 4.45GYY315 pKa = 10.27WRR317 pKa = 11.84TSVPAADD324 pKa = 3.81VAALPDD330 pKa = 4.03GAQLVATVTNALGRR344 pKa = 11.84EE345 pKa = 4.26AGDD348 pKa = 3.66SATLTVNSSLPSLTVDD364 pKa = 4.31PFATDD369 pKa = 3.22NVLSDD374 pKa = 5.16AEE376 pKa = 4.17AQSAQTLSGTTTGVEE391 pKa = 3.68AGQAVTVTLNEE402 pKa = 3.81ITYY405 pKa = 8.07STTVAADD412 pKa = 3.71GSFSLSVPSADD423 pKa = 5.09LLALYY428 pKa = 10.32SGQTTIAARR437 pKa = 11.84AANLAGQEE445 pKa = 4.37TQGSEE450 pKa = 3.55ILTVDD455 pKa = 4.77FATARR460 pKa = 11.84LVVDD464 pKa = 4.72PVAGDD469 pKa = 3.5NYY471 pKa = 10.3INASEE476 pKa = 4.05SSYY479 pKa = 11.35LVLSGTTVDD488 pKa = 3.83VPAGSWVVITFNDD501 pKa = 4.06RR502 pKa = 11.84IFRR505 pKa = 11.84AEE507 pKa = 3.96VEE509 pKa = 4.46ADD511 pKa = 4.04GSWSTYY517 pKa = 10.53VSDD520 pKa = 4.46LVVNSFPDD528 pKa = 3.57GQYY531 pKa = 10.46PITFSAPGSAPVTLDD546 pKa = 2.86VGLYY550 pKa = 9.84IDD552 pKa = 4.82NQAQLNVQIDD562 pKa = 4.02TLFGDD567 pKa = 4.79DD568 pKa = 3.8VLTPGEE574 pKa = 4.15TLQDD578 pKa = 3.49QVLTGTTGITGAGQTISAEE597 pKa = 4.26LEE599 pKa = 4.48GVTHH603 pKa = 6.89TGSVDD608 pKa = 3.2NDD610 pKa = 4.08GNWTITVPAAALQALQNPNGYY631 pKa = 10.36VYY633 pKa = 10.92VQAADD638 pKa = 3.58LAGNSIAVGRR648 pKa = 11.84SFEE651 pKa = 4.91LVFSPATLTIDD662 pKa = 5.46PITGDD667 pKa = 3.91NILTAAEE674 pKa = 4.2TATPLTVSGVSYY686 pKa = 10.23RR687 pKa = 11.84VPEE690 pKa = 4.32GVTITVTLNGKK701 pKa = 9.56EE702 pKa = 4.13YY703 pKa = 7.83TTTAPGNDD711 pKa = 3.24TWSVQVPAADD721 pKa = 3.97LAGLTDD727 pKa = 3.91GVYY730 pKa = 9.55TVTAATINIDD740 pKa = 3.91LEE742 pKa = 4.51PVTTQQDD749 pKa = 3.89LTVDD753 pKa = 4.55LAPAPAFSVDD763 pKa = 3.88PVTGDD768 pKa = 3.9DD769 pKa = 3.4ILYY772 pKa = 10.17VDD774 pKa = 4.11EE775 pKa = 4.55TSGEE779 pKa = 3.96IVITGRR785 pKa = 11.84APVSAVGGSVTVTAFDD801 pKa = 4.04DD802 pKa = 3.54PAYY805 pKa = 10.44RR806 pKa = 11.84YY807 pKa = 7.59TGTVAADD814 pKa = 3.79GTWQVTVPEE823 pKa = 4.39YY824 pKa = 11.36NFGGTSNGVYY834 pKa = 9.85TLVAEE839 pKa = 4.71LTAPDD844 pKa = 3.91GTTTTTTHH852 pKa = 7.66DD853 pKa = 2.94ITINVWPFSGDD864 pKa = 3.29FYY866 pKa = 11.49FDD868 pKa = 4.79DD869 pKa = 4.02FTGDD873 pKa = 3.23STLTAEE879 pKa = 5.45EE880 pKa = 4.22RR881 pKa = 11.84TQDD884 pKa = 3.45QVFSGTFASEE894 pKa = 4.09EE895 pKa = 4.09NDD897 pKa = 3.29VSGLGITVYY906 pKa = 9.7MHH908 pKa = 7.42DD909 pKa = 4.17NDD911 pKa = 4.37GQTFAYY917 pKa = 9.41TGTTDD922 pKa = 3.67GNGRR926 pKa = 11.84WSVTIPAADD935 pKa = 3.64LQQLSEE941 pKa = 4.02GWSYY945 pKa = 7.65MTATATDD952 pKa = 3.42AAGNGASSSRR962 pKa = 11.84EE963 pKa = 3.22ADD965 pKa = 3.78FAVLPATGGTITVNPIAGDD984 pKa = 3.62NVLTADD990 pKa = 4.93EE991 pKa = 4.81ISDD994 pKa = 3.69GLTISGSSTYY1004 pKa = 9.8MGEE1007 pKa = 4.17RR1008 pKa = 11.84YY1009 pKa = 7.47TQIEE1013 pKa = 4.17VEE1015 pKa = 4.15INGTLRR1021 pKa = 11.84IYY1023 pKa = 11.25ADD1025 pKa = 4.12YY1026 pKa = 10.95EE1027 pKa = 4.49SDD1029 pKa = 2.92WSVTFSRR1036 pKa = 11.84EE1037 pKa = 4.0FIATLPDD1044 pKa = 3.67GPLTVTASGYY1054 pKa = 10.15DD1055 pKa = 3.34LSGNRR1060 pKa = 11.84VTTTATLEE1068 pKa = 4.25VNLSDD1073 pKa = 4.91VSTLTLDD1080 pKa = 3.63PVTGDD1085 pKa = 3.92DD1086 pKa = 4.89VISADD1091 pKa = 3.57EE1092 pKa = 4.33AGGEE1096 pKa = 4.07IVITGRR1102 pKa = 11.84APLSTQGGTVTVTAFDD1118 pKa = 4.13DD1119 pKa = 3.53PAYY1122 pKa = 10.4RR1123 pKa = 11.84YY1124 pKa = 6.88TATVEE1129 pKa = 4.89EE1130 pKa = 5.09GGTWQATVPAGAFSGRR1146 pKa = 11.84ANGVYY1151 pKa = 9.79TLVAEE1156 pKa = 4.71LTAPDD1161 pKa = 3.88GTVTTLNHH1169 pKa = 6.92DD1170 pKa = 3.68VTSNQLPFSGGFAFYY1185 pKa = 11.02VFAGDD1190 pKa = 3.78DD1191 pKa = 3.45VLNAEE1196 pKa = 4.87EE1197 pKa = 4.0QAQDD1201 pKa = 3.46QILEE1205 pKa = 4.36GEE1207 pKa = 4.52VFSDD1211 pKa = 4.67DD1212 pKa = 3.79GGNVAGLAVTIQMTTFDD1229 pKa = 4.33GQQFTYY1235 pKa = 8.05TTTTGEE1241 pKa = 3.69NGYY1244 pKa = 8.39WQVTLPSSDD1253 pKa = 4.43LEE1255 pKa = 3.95QLRR1258 pKa = 11.84NGWVNLVGTVADD1270 pKa = 4.16GAGNTLSNEE1279 pKa = 4.15SQFTVLAGIGTLTINPIAGDD1299 pKa = 3.68NVLTADD1305 pKa = 4.31EE1306 pKa = 4.73IGNGLVVSGSGEE1318 pKa = 4.1FLDD1321 pKa = 4.23MPLSLEE1327 pKa = 3.99VTINGVSQGFSDD1339 pKa = 4.83YY1340 pKa = 11.38SSDD1343 pKa = 2.81WSVTFTPQQLAGFPDD1358 pKa = 4.56GPLTVTVTAYY1368 pKa = 10.36DD1369 pKa = 3.37IEE1371 pKa = 4.89FNLITATSTLTVDD1384 pKa = 5.3LGDD1387 pKa = 4.16LQPLTFNPFGGDD1399 pKa = 3.55DD1400 pKa = 3.7NYY1402 pKa = 11.42LDD1404 pKa = 4.19EE1405 pKa = 6.73DD1406 pKa = 4.18EE1407 pKa = 5.81LEE1409 pKa = 4.27SDD1411 pKa = 4.1QLLSGQAGNLAAGSIVTLTLGEE1433 pKa = 4.01FTYY1436 pKa = 10.56QAEE1439 pKa = 4.42VQADD1443 pKa = 3.89HH1444 pKa = 6.48SWSVLIPSADD1454 pKa = 3.86LQTLPNDD1461 pKa = 3.22ATTGISATVTDD1472 pKa = 4.25ADD1474 pKa = 4.36GNLLAAGSHH1483 pKa = 5.26TFLLYY1488 pKa = 8.6RR1489 pKa = 11.84TPGEE1493 pKa = 4.1LNIDD1497 pKa = 3.73TLAGNGILTADD1508 pKa = 4.12EE1509 pKa = 4.25ATVPLVISGTITNGRR1524 pKa = 11.84VGAAVTVDD1532 pKa = 4.07LNGHH1536 pKa = 7.27LYY1538 pKa = 8.17QTTVTASDD1546 pKa = 4.28GSWQVSVPVEE1556 pKa = 4.34DD1557 pKa = 4.74LAQLGNTEE1565 pKa = 4.25YY1566 pKa = 10.01WVSATSTDD1574 pKa = 3.83LSGNEE1579 pKa = 3.75INGLNLLVVDD1589 pKa = 5.45LYY1591 pKa = 11.84VPVITLDD1598 pKa = 4.31PIAGDD1603 pKa = 3.86NIINAAEE1610 pKa = 3.92AAAGVVLSGQSSEE1623 pKa = 4.17AGGVVSVQLNGVAYY1637 pKa = 9.05EE1638 pKa = 4.16AGVDD1642 pKa = 4.83EE1643 pKa = 4.73EE1644 pKa = 5.53GSWQLALPNSALAEE1658 pKa = 4.33LEE1660 pKa = 4.07DD1661 pKa = 3.94GRR1663 pKa = 11.84YY1664 pKa = 7.65TLTLSQTGFNGQTTTVTEE1682 pKa = 4.2SLLLDD1687 pKa = 4.37ADD1689 pKa = 3.96PANRR1693 pKa = 11.84PVLRR1697 pKa = 11.84IHH1699 pKa = 7.15TISHH1703 pKa = 6.77DD1704 pKa = 3.65NVLNGAEE1711 pKa = 4.21LQSDD1715 pKa = 3.85QIITGSSEE1723 pKa = 3.94NVEE1726 pKa = 4.29SGQTLSLKK1734 pKa = 10.42IGGVTYY1740 pKa = 7.72TTQVQPGGGWSLIVPAEE1757 pKa = 4.11DD1758 pKa = 3.93LAALGDD1764 pKa = 3.86GGLTLRR1770 pKa = 11.84AGVTDD1775 pKa = 3.93ASGNRR1780 pKa = 11.84ATASRR1785 pKa = 11.84NLTIKK1790 pKa = 10.26SDD1792 pKa = 3.85RR1793 pKa = 11.84DD1794 pKa = 3.58GLSIDD1799 pKa = 4.35PVTGDD1804 pKa = 3.77NLINAADD1811 pKa = 3.8AEE1813 pKa = 4.55GAITLTGHH1821 pKa = 6.04TDD1823 pKa = 3.71GVRR1826 pKa = 11.84HH1827 pKa = 5.99GATVRR1832 pKa = 11.84LTLNGEE1838 pKa = 4.03HH1839 pKa = 5.76YY1840 pKa = 7.81TAKK1843 pKa = 10.3VGKK1846 pKa = 9.86DD1847 pKa = 3.57GSWSAKK1853 pKa = 9.74VPAADD1858 pKa = 3.35VAALEE1863 pKa = 4.61EE1864 pKa = 5.01GSATLTACVKK1874 pKa = 9.85SACGQMLYY1882 pKa = 10.91AKK1884 pKa = 10.34AVLTVDD1890 pKa = 3.31TRR1892 pKa = 11.84APEE1895 pKa = 4.13LALLPVTGDD1904 pKa = 3.56NLISLQEE1911 pKa = 4.04AQAGFALTGSAGIAAAGLLVLVVLNGVEE1939 pKa = 4.19YY1940 pKa = 10.2QGKK1943 pKa = 8.51VQASGDD1949 pKa = 3.47WTVAIPAGTLADD1961 pKa = 4.17QPAGDD1966 pKa = 4.01YY1967 pKa = 10.28PLTVSLTDD1975 pKa = 3.26TAGNLTSLTSVVTLEE1990 pKa = 4.3AAPDD1994 pKa = 3.56ADD1996 pKa = 3.75ATEE1999 pKa = 4.41TAALTVTSQTVAASADD2015 pKa = 3.7STLTSALAVPDD2026 pKa = 4.19TATDD2030 pKa = 3.71TADD2033 pKa = 2.94GTYY2036 pKa = 10.45AIGGQTLTLTEE2047 pKa = 4.37SGGEE2051 pKa = 3.91ALGGSGNDD2059 pKa = 3.53TILLNTLDD2067 pKa = 4.37FLHH2070 pKa = 7.2IDD2072 pKa = 4.04GGSGTDD2078 pKa = 3.35TLLLAGSDD2086 pKa = 3.36QHH2088 pKa = 8.28LDD2090 pKa = 3.34LTTLGLKK2097 pKa = 9.77IEE2099 pKa = 4.97NIDD2102 pKa = 4.76IFDD2105 pKa = 4.87LGNGSNSLTLGLHH2118 pKa = 5.09EE2119 pKa = 5.32AEE2121 pKa = 4.65TVRR2124 pKa = 11.84DD2125 pKa = 3.94TPEE2128 pKa = 3.39EE2129 pKa = 4.02SLFIRR2134 pKa = 11.84GQGGSQLTLAGDD2146 pKa = 4.13NTWEE2150 pKa = 3.97TSGQRR2155 pKa = 11.84EE2156 pKa = 4.21VGGLLFDD2163 pKa = 4.44VYY2165 pKa = 10.61HH2166 pKa = 5.88ATGLEE2171 pKa = 4.21SADD2174 pKa = 4.35LLVQQGILVQQGG2186 pKa = 2.84
Molecular weight: 225.74 kDa
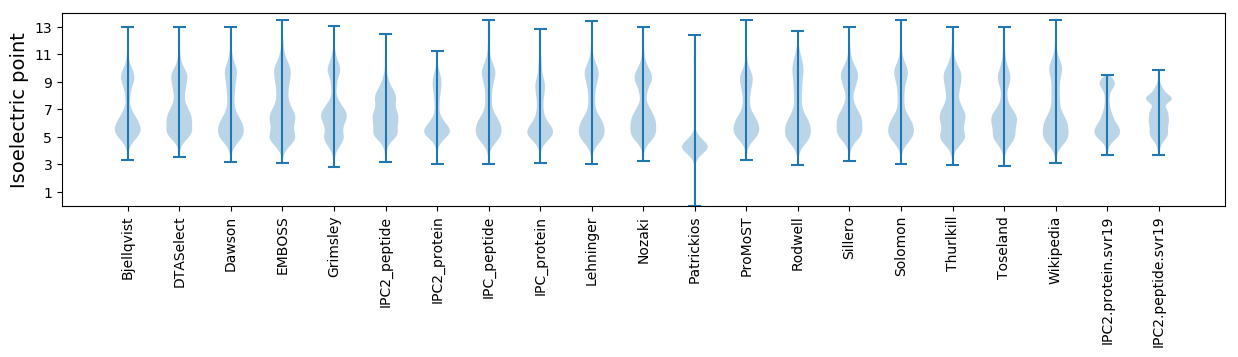
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2N5ELK9|A0A2N5ELK9_9GAMM FMN-binding protein MioC OS=Chimaeribacter arupi OX=2060066 GN=CYR34_13685 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.6RR12 pKa = 11.84NRR14 pKa = 11.84SHH16 pKa = 7.16GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.25GRR39 pKa = 11.84ARR41 pKa = 11.84LSVSKK46 pKa = 10.99
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.6RR12 pKa = 11.84NRR14 pKa = 11.84SHH16 pKa = 7.16GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.25GRR39 pKa = 11.84ARR41 pKa = 11.84LSVSKK46 pKa = 10.99
Molecular weight: 5.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1358501 |
15 |
6646 |
326.6 |
35.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.823 ± 0.055 | 1.017 ± 0.014 |
5.151 ± 0.038 | 5.344 ± 0.037 |
3.744 ± 0.028 | 7.739 ± 0.037 |
2.388 ± 0.022 | 5.206 ± 0.035 |
3.541 ± 0.038 | 11.169 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.625 ± 0.024 | 3.406 ± 0.031 |
4.924 ± 0.03 | 4.633 ± 0.037 |
5.901 ± 0.047 | 5.557 ± 0.032 |
5.504 ± 0.065 | 7.129 ± 0.035 |
1.416 ± 0.016 | 2.782 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
