
Bat polyomavirus 6d
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; unclassified Polyomaviridae
Average proteome isoelectric point is 5.5
Get precalculated fractions of proteins
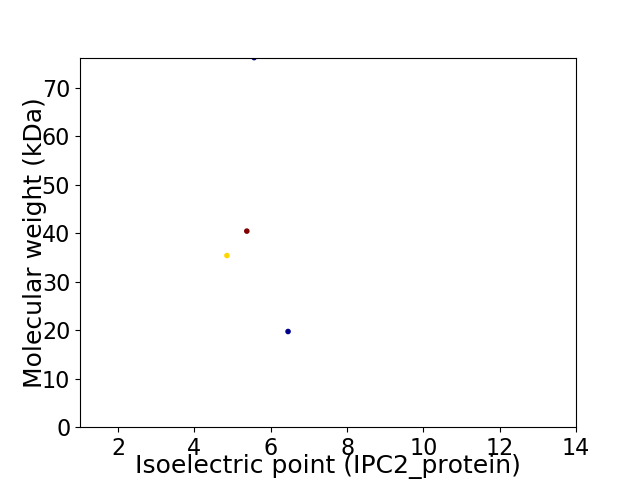
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D5ZYQ7|A0A0D5ZYQ7_9POLY Small t antigen OS=Bat polyomavirus 6d OX=1623686 PE=4 SV=1
MM1 pKa = 7.31GALLAVLAEE10 pKa = 4.54VIDD13 pKa = 4.36LSAVTGISVEE23 pKa = 4.37GILAGEE29 pKa = 4.48AFATAEE35 pKa = 3.94VLAAQIEE42 pKa = 4.75NITLLEE48 pKa = 4.19GLTAAEE54 pKa = 4.39AVAEE58 pKa = 4.11LGYY61 pKa = 9.24TVDD64 pKa = 3.54VFNALASVTEE74 pKa = 4.05NFPEE78 pKa = 4.45VFTLLAAGEE87 pKa = 4.27VAANSSLVLSAAVAAATYY105 pKa = 8.63PYY107 pKa = 10.09SYY109 pKa = 9.28THH111 pKa = 6.44EE112 pKa = 4.19VPIANLNNMALQVWQPEE129 pKa = 3.73IDD131 pKa = 3.34ILFPGISSLARR142 pKa = 11.84FLNYY146 pKa = 9.33IDD148 pKa = 3.62PTNWAGDD155 pKa = 3.49LFRR158 pKa = 11.84AVGRR162 pKa = 11.84YY163 pKa = 7.21FWEE166 pKa = 4.06TAQRR170 pKa = 11.84SGRR173 pKa = 11.84NLLEE177 pKa = 3.66QEE179 pKa = 3.65IRR181 pKa = 11.84AVGQRR186 pKa = 11.84AAGGFSEE193 pKa = 4.2TLARR197 pKa = 11.84YY198 pKa = 9.21FEE200 pKa = 4.05NARR203 pKa = 11.84WAIRR207 pKa = 11.84HH208 pKa = 5.4YY209 pKa = 10.56PADD212 pKa = 3.66LYY214 pKa = 11.58GRR216 pKa = 11.84LQQYY220 pKa = 7.71YY221 pKa = 10.04TDD223 pKa = 5.11LPPLNPAQLRR233 pKa = 11.84EE234 pKa = 3.78LSRR237 pKa = 11.84RR238 pKa = 11.84VGGPDD243 pKa = 3.42QPYY246 pKa = 10.53KK247 pKa = 10.94LYY249 pKa = 10.67DD250 pKa = 3.51SHH252 pKa = 7.94VKK254 pKa = 10.17SAEE257 pKa = 4.05FVTKK261 pKa = 10.51SPPPGGAHH269 pKa = 6.18QRR271 pKa = 11.84TTPDD275 pKa = 2.65WLLPLILGLYY285 pKa = 10.37GDD287 pKa = 5.25ISPSWADD294 pKa = 3.23TLEE297 pKa = 4.06EE298 pKa = 4.48LEE300 pKa = 4.54EE301 pKa = 4.54EE302 pKa = 4.01EE303 pKa = 5.96DD304 pKa = 4.02GPKK307 pKa = 9.87KK308 pKa = 10.26KK309 pKa = 10.08KK310 pKa = 9.9RR311 pKa = 11.84ASQRR315 pKa = 11.84SSRR318 pKa = 11.84SAKK321 pKa = 9.27TNNN324 pKa = 3.08
MM1 pKa = 7.31GALLAVLAEE10 pKa = 4.54VIDD13 pKa = 4.36LSAVTGISVEE23 pKa = 4.37GILAGEE29 pKa = 4.48AFATAEE35 pKa = 3.94VLAAQIEE42 pKa = 4.75NITLLEE48 pKa = 4.19GLTAAEE54 pKa = 4.39AVAEE58 pKa = 4.11LGYY61 pKa = 9.24TVDD64 pKa = 3.54VFNALASVTEE74 pKa = 4.05NFPEE78 pKa = 4.45VFTLLAAGEE87 pKa = 4.27VAANSSLVLSAAVAAATYY105 pKa = 8.63PYY107 pKa = 10.09SYY109 pKa = 9.28THH111 pKa = 6.44EE112 pKa = 4.19VPIANLNNMALQVWQPEE129 pKa = 3.73IDD131 pKa = 3.34ILFPGISSLARR142 pKa = 11.84FLNYY146 pKa = 9.33IDD148 pKa = 3.62PTNWAGDD155 pKa = 3.49LFRR158 pKa = 11.84AVGRR162 pKa = 11.84YY163 pKa = 7.21FWEE166 pKa = 4.06TAQRR170 pKa = 11.84SGRR173 pKa = 11.84NLLEE177 pKa = 3.66QEE179 pKa = 3.65IRR181 pKa = 11.84AVGQRR186 pKa = 11.84AAGGFSEE193 pKa = 4.2TLARR197 pKa = 11.84YY198 pKa = 9.21FEE200 pKa = 4.05NARR203 pKa = 11.84WAIRR207 pKa = 11.84HH208 pKa = 5.4YY209 pKa = 10.56PADD212 pKa = 3.66LYY214 pKa = 11.58GRR216 pKa = 11.84LQQYY220 pKa = 7.71YY221 pKa = 10.04TDD223 pKa = 5.11LPPLNPAQLRR233 pKa = 11.84EE234 pKa = 3.78LSRR237 pKa = 11.84RR238 pKa = 11.84VGGPDD243 pKa = 3.42QPYY246 pKa = 10.53KK247 pKa = 10.94LYY249 pKa = 10.67DD250 pKa = 3.51SHH252 pKa = 7.94VKK254 pKa = 10.17SAEE257 pKa = 4.05FVTKK261 pKa = 10.51SPPPGGAHH269 pKa = 6.18QRR271 pKa = 11.84TTPDD275 pKa = 2.65WLLPLILGLYY285 pKa = 10.37GDD287 pKa = 5.25ISPSWADD294 pKa = 3.23TLEE297 pKa = 4.06EE298 pKa = 4.48LEE300 pKa = 4.54EE301 pKa = 4.54EE302 pKa = 4.01EE303 pKa = 5.96DD304 pKa = 4.02GPKK307 pKa = 9.87KK308 pKa = 10.26KK309 pKa = 10.08KK310 pKa = 9.9RR311 pKa = 11.84ASQRR315 pKa = 11.84SSRR318 pKa = 11.84SAKK321 pKa = 9.27TNNN324 pKa = 3.08
Molecular weight: 35.42 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D5ZYQ7|A0A0D5ZYQ7_9POLY Small t antigen OS=Bat polyomavirus 6d OX=1623686 PE=4 SV=1
MM1 pKa = 7.77DD2 pKa = 5.3HH3 pKa = 7.29LLTRR7 pKa = 11.84EE8 pKa = 3.66EE9 pKa = 4.76SIRR12 pKa = 11.84LMQLLEE18 pKa = 4.24LPMDD22 pKa = 3.65EE23 pKa = 4.77FGNFNAMRR31 pKa = 11.84SQFHH35 pKa = 5.6KK36 pKa = 10.25QIKK39 pKa = 9.53KK40 pKa = 6.86MHH42 pKa = 6.94PDD44 pKa = 2.82KK45 pKa = 11.35GGNPEE50 pKa = 3.83QAKK53 pKa = 10.2EE54 pKa = 4.03LISLYY59 pKa = 10.69KK60 pKa = 10.38KK61 pKa = 10.66LEE63 pKa = 4.13EE64 pKa = 5.23KK65 pKa = 10.38ISPLQPDD72 pKa = 3.25SDD74 pKa = 3.82ATTTEE79 pKa = 3.71QVYY82 pKa = 9.71EE83 pKa = 3.87QGQFFLYY90 pKa = 10.6LKK92 pKa = 10.57DD93 pKa = 3.18WVSCNRR99 pKa = 11.84GEE101 pKa = 4.54SSCACLFCMLKK112 pKa = 10.7ANHH115 pKa = 6.24EE116 pKa = 4.02KK117 pKa = 10.46RR118 pKa = 11.84KK119 pKa = 10.0RR120 pKa = 11.84EE121 pKa = 3.83RR122 pKa = 11.84KK123 pKa = 8.42HH124 pKa = 6.19NVWGHH129 pKa = 5.65CFCFQCYY136 pKa = 7.7ITWFGLEE143 pKa = 4.33HH144 pKa = 7.39SWMDD148 pKa = 3.13WLSWRR153 pKa = 11.84NVIANTPYY161 pKa = 10.73GCLNII166 pKa = 4.6
MM1 pKa = 7.77DD2 pKa = 5.3HH3 pKa = 7.29LLTRR7 pKa = 11.84EE8 pKa = 3.66EE9 pKa = 4.76SIRR12 pKa = 11.84LMQLLEE18 pKa = 4.24LPMDD22 pKa = 3.65EE23 pKa = 4.77FGNFNAMRR31 pKa = 11.84SQFHH35 pKa = 5.6KK36 pKa = 10.25QIKK39 pKa = 9.53KK40 pKa = 6.86MHH42 pKa = 6.94PDD44 pKa = 2.82KK45 pKa = 11.35GGNPEE50 pKa = 3.83QAKK53 pKa = 10.2EE54 pKa = 4.03LISLYY59 pKa = 10.69KK60 pKa = 10.38KK61 pKa = 10.66LEE63 pKa = 4.13EE64 pKa = 5.23KK65 pKa = 10.38ISPLQPDD72 pKa = 3.25SDD74 pKa = 3.82ATTTEE79 pKa = 3.71QVYY82 pKa = 9.71EE83 pKa = 3.87QGQFFLYY90 pKa = 10.6LKK92 pKa = 10.57DD93 pKa = 3.18WVSCNRR99 pKa = 11.84GEE101 pKa = 4.54SSCACLFCMLKK112 pKa = 10.7ANHH115 pKa = 6.24EE116 pKa = 4.02KK117 pKa = 10.46RR118 pKa = 11.84KK119 pKa = 10.0RR120 pKa = 11.84EE121 pKa = 3.83RR122 pKa = 11.84KK123 pKa = 8.42HH124 pKa = 6.19NVWGHH129 pKa = 5.65CFCFQCYY136 pKa = 7.7ITWFGLEE143 pKa = 4.33HH144 pKa = 7.39SWMDD148 pKa = 3.13WLSWRR153 pKa = 11.84NVIANTPYY161 pKa = 10.73GCLNII166 pKa = 4.6
Molecular weight: 19.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1518 |
166 |
661 |
379.5 |
42.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.588 ± 1.983 | 2.306 ± 0.886 |
5.468 ± 0.609 | 7.642 ± 0.388 |
4.875 ± 0.644 | 5.863 ± 0.977 |
1.845 ± 0.463 | 4.677 ± 0.175 |
6.522 ± 1.349 | 10.738 ± 0.498 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.174 ± 0.535 | 4.875 ± 0.33 |
5.138 ± 0.83 | 4.414 ± 0.456 |
4.611 ± 0.299 | 5.863 ± 0.225 |
6.258 ± 0.623 | 5.402 ± 0.707 |
1.383 ± 0.486 | 3.36 ± 0.317 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
