
Beihai tombus-like virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 9.06
Get precalculated fractions of proteins
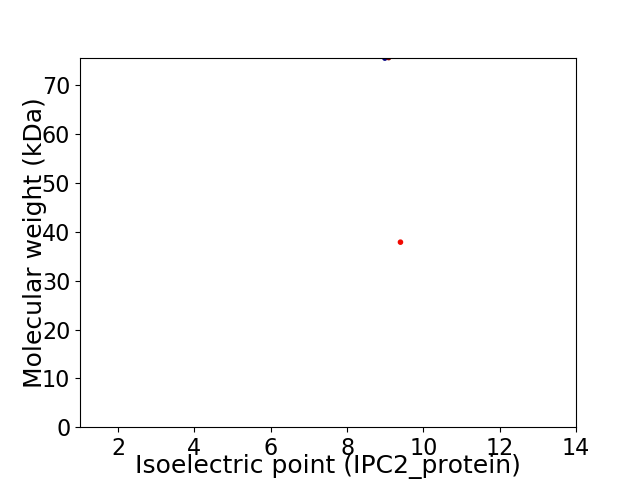
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFQ7|A0A1L3KFQ7_9VIRU Uncharacterized protein OS=Beihai tombus-like virus 2 OX=1922723 PE=4 SV=1
MM1 pKa = 7.56SISDD5 pKa = 3.51VLQIQKK11 pKa = 10.73GVTCLSATIPRR22 pKa = 11.84QVRR25 pKa = 11.84RR26 pKa = 11.84LPRR29 pKa = 11.84VFYY32 pKa = 10.29TIAYY36 pKa = 8.24ANNFRR41 pKa = 11.84IPNNDD46 pKa = 3.51LVTKK50 pKa = 9.62LHH52 pKa = 6.29AFVEE56 pKa = 4.96RR57 pKa = 11.84VCLVRR62 pKa = 11.84NSEE65 pKa = 4.29GNLVATPRR73 pKa = 11.84PWDD76 pKa = 3.36VLMSEE81 pKa = 5.2DD82 pKa = 3.56VDD84 pKa = 3.9YY85 pKa = 11.51DD86 pKa = 3.43QAEE89 pKa = 4.02QLFIDD94 pKa = 3.7MTAPVVEE101 pKa = 4.3ALSRR105 pKa = 11.84EE106 pKa = 4.49YY107 pKa = 9.43ITAGGVDD114 pKa = 3.53QPITYY119 pKa = 10.03DD120 pKa = 3.07QFLNHH125 pKa = 5.86YY126 pKa = 8.95VGSKK130 pKa = 9.94RR131 pKa = 11.84KK132 pKa = 9.54IYY134 pKa = 9.44EE135 pKa = 3.64QAIEE139 pKa = 4.25SLRR142 pKa = 11.84DD143 pKa = 3.09IPFQDD148 pKa = 3.3KK149 pKa = 10.64RR150 pKa = 11.84DD151 pKa = 3.57GSVEE155 pKa = 3.78IFIKK159 pKa = 10.57PEE161 pKa = 3.62YY162 pKa = 9.19LKK164 pKa = 10.77PGGVPRR170 pKa = 11.84VISPRR175 pKa = 11.84HH176 pKa = 5.04PRR178 pKa = 11.84YY179 pKa = 9.22HH180 pKa = 5.78ATIGLYY186 pKa = 9.86IKK188 pKa = 10.52AIEE191 pKa = 4.1DD192 pKa = 3.67KK193 pKa = 10.15IYY195 pKa = 11.33NSIDD199 pKa = 3.64TMWDD203 pKa = 2.9PTGQFKK209 pKa = 9.02TVAKK213 pKa = 9.93KK214 pKa = 9.86MDD216 pKa = 3.89MIQRR220 pKa = 11.84AEE222 pKa = 4.44EE223 pKa = 4.06LNKK226 pKa = 9.23MWSSYY231 pKa = 9.58GRR233 pKa = 11.84PRR235 pKa = 11.84AVGFDD240 pKa = 3.24AKK242 pKa = 10.8RR243 pKa = 11.84FDD245 pKa = 3.32QHH247 pKa = 6.72INSWLLKK254 pKa = 9.27HH255 pKa = 6.59LEE257 pKa = 3.78HH258 pKa = 7.4KK259 pKa = 10.16IYY261 pKa = 10.91KK262 pKa = 9.1KK263 pKa = 10.11VCHH266 pKa = 5.59NTSPEE271 pKa = 4.08HH272 pKa = 6.18EE273 pKa = 4.48VTLSRR278 pKa = 11.84LLEE281 pKa = 4.09LQQTNIIKK289 pKa = 10.8VGDD292 pKa = 3.89RR293 pKa = 11.84KK294 pKa = 11.2DD295 pKa = 3.14MARR298 pKa = 11.84YY299 pKa = 6.8TFEE302 pKa = 3.87VDD304 pKa = 3.3GVRR307 pKa = 11.84MSGDD311 pKa = 3.33MNTSLGNVTIMCCLMWLFRR330 pKa = 11.84EE331 pKa = 4.77EE332 pKa = 3.9YY333 pKa = 10.6HH334 pKa = 7.44FDD336 pKa = 3.55FKK338 pKa = 10.91MLNDD342 pKa = 4.02GDD344 pKa = 4.67DD345 pKa = 3.77LVIVCDD351 pKa = 3.56RR352 pKa = 11.84VTAKK356 pKa = 10.53AIKK359 pKa = 10.18DD360 pKa = 3.5KK361 pKa = 11.23LPSFFLKK368 pKa = 10.67FGIEE372 pKa = 3.98MEE374 pKa = 4.34FEE376 pKa = 4.47GLFHH380 pKa = 6.38TLEE383 pKa = 5.22AIEE386 pKa = 4.71FCQAHH391 pKa = 6.21PVQYY395 pKa = 10.91DD396 pKa = 3.25VGKK399 pKa = 9.71WIMVPNPSKK408 pKa = 10.93RR409 pKa = 11.84IFSDD413 pKa = 3.64LLSEE417 pKa = 4.65KK418 pKa = 9.98PVEE421 pKa = 4.06SRR423 pKa = 11.84KK424 pKa = 9.88IYY426 pKa = 10.41SSWLGAVGKK435 pKa = 9.23CGCAMSPGVPILQEE449 pKa = 4.06FYY451 pKa = 11.23DD452 pKa = 4.22WILRR456 pKa = 11.84TPGVKK461 pKa = 9.54PWIPKK466 pKa = 9.18EE467 pKa = 3.34GSYY470 pKa = 10.34YY471 pKa = 10.55YY472 pKa = 10.61KK473 pKa = 9.98YY474 pKa = 10.74GYY476 pKa = 10.11RR477 pKa = 11.84KK478 pKa = 9.88VIEE481 pKa = 4.14GAKK484 pKa = 9.08YY485 pKa = 10.33KK486 pKa = 10.12PVSEE490 pKa = 4.15IARR493 pKa = 11.84ISFFKK498 pKa = 10.5AYY500 pKa = 9.96NIPPSEE506 pKa = 4.02QIMLEE511 pKa = 3.85NMFRR515 pKa = 11.84SLPSPSRR522 pKa = 11.84EE523 pKa = 3.6ILEE526 pKa = 4.26EE527 pKa = 3.94SSRR530 pKa = 11.84PAHH533 pKa = 5.55TLPLEE538 pKa = 4.23MFALPTYY545 pKa = 9.97FKK547 pKa = 10.71KK548 pKa = 10.98FNWEE552 pKa = 3.38EE553 pKa = 3.74TDD555 pKa = 4.36RR556 pKa = 11.84MNRR559 pKa = 11.84WEE561 pKa = 4.28LDD563 pKa = 2.93RR564 pKa = 11.84GLFLVKK570 pKa = 10.26NKK572 pKa = 9.51RR573 pKa = 11.84RR574 pKa = 11.84AQDD577 pKa = 3.06KK578 pKa = 10.42GKK580 pKa = 11.32SNLTVKK586 pKa = 10.1NVKK589 pKa = 9.72KK590 pKa = 10.75DD591 pKa = 3.85PIRR594 pKa = 11.84FRR596 pKa = 11.84KK597 pKa = 9.02PKK599 pKa = 9.08RR600 pKa = 11.84SQRR603 pKa = 11.84RR604 pKa = 11.84RR605 pKa = 11.84ARR607 pKa = 11.84YY608 pKa = 6.61TSKK611 pKa = 10.35HH612 pKa = 5.26CRR614 pKa = 11.84HH615 pKa = 6.48RR616 pKa = 11.84SAKK619 pKa = 8.8TRR621 pKa = 11.84SAVTNLTKK629 pKa = 10.4SSRR632 pKa = 11.84QKK634 pKa = 10.32RR635 pKa = 11.84RR636 pKa = 11.84RR637 pKa = 11.84SAYY640 pKa = 8.48RR641 pKa = 11.84FRR643 pKa = 11.84YY644 pKa = 9.59RR645 pKa = 11.84SAA647 pKa = 3.46
MM1 pKa = 7.56SISDD5 pKa = 3.51VLQIQKK11 pKa = 10.73GVTCLSATIPRR22 pKa = 11.84QVRR25 pKa = 11.84RR26 pKa = 11.84LPRR29 pKa = 11.84VFYY32 pKa = 10.29TIAYY36 pKa = 8.24ANNFRR41 pKa = 11.84IPNNDD46 pKa = 3.51LVTKK50 pKa = 9.62LHH52 pKa = 6.29AFVEE56 pKa = 4.96RR57 pKa = 11.84VCLVRR62 pKa = 11.84NSEE65 pKa = 4.29GNLVATPRR73 pKa = 11.84PWDD76 pKa = 3.36VLMSEE81 pKa = 5.2DD82 pKa = 3.56VDD84 pKa = 3.9YY85 pKa = 11.51DD86 pKa = 3.43QAEE89 pKa = 4.02QLFIDD94 pKa = 3.7MTAPVVEE101 pKa = 4.3ALSRR105 pKa = 11.84EE106 pKa = 4.49YY107 pKa = 9.43ITAGGVDD114 pKa = 3.53QPITYY119 pKa = 10.03DD120 pKa = 3.07QFLNHH125 pKa = 5.86YY126 pKa = 8.95VGSKK130 pKa = 9.94RR131 pKa = 11.84KK132 pKa = 9.54IYY134 pKa = 9.44EE135 pKa = 3.64QAIEE139 pKa = 4.25SLRR142 pKa = 11.84DD143 pKa = 3.09IPFQDD148 pKa = 3.3KK149 pKa = 10.64RR150 pKa = 11.84DD151 pKa = 3.57GSVEE155 pKa = 3.78IFIKK159 pKa = 10.57PEE161 pKa = 3.62YY162 pKa = 9.19LKK164 pKa = 10.77PGGVPRR170 pKa = 11.84VISPRR175 pKa = 11.84HH176 pKa = 5.04PRR178 pKa = 11.84YY179 pKa = 9.22HH180 pKa = 5.78ATIGLYY186 pKa = 9.86IKK188 pKa = 10.52AIEE191 pKa = 4.1DD192 pKa = 3.67KK193 pKa = 10.15IYY195 pKa = 11.33NSIDD199 pKa = 3.64TMWDD203 pKa = 2.9PTGQFKK209 pKa = 9.02TVAKK213 pKa = 9.93KK214 pKa = 9.86MDD216 pKa = 3.89MIQRR220 pKa = 11.84AEE222 pKa = 4.44EE223 pKa = 4.06LNKK226 pKa = 9.23MWSSYY231 pKa = 9.58GRR233 pKa = 11.84PRR235 pKa = 11.84AVGFDD240 pKa = 3.24AKK242 pKa = 10.8RR243 pKa = 11.84FDD245 pKa = 3.32QHH247 pKa = 6.72INSWLLKK254 pKa = 9.27HH255 pKa = 6.59LEE257 pKa = 3.78HH258 pKa = 7.4KK259 pKa = 10.16IYY261 pKa = 10.91KK262 pKa = 9.1KK263 pKa = 10.11VCHH266 pKa = 5.59NTSPEE271 pKa = 4.08HH272 pKa = 6.18EE273 pKa = 4.48VTLSRR278 pKa = 11.84LLEE281 pKa = 4.09LQQTNIIKK289 pKa = 10.8VGDD292 pKa = 3.89RR293 pKa = 11.84KK294 pKa = 11.2DD295 pKa = 3.14MARR298 pKa = 11.84YY299 pKa = 6.8TFEE302 pKa = 3.87VDD304 pKa = 3.3GVRR307 pKa = 11.84MSGDD311 pKa = 3.33MNTSLGNVTIMCCLMWLFRR330 pKa = 11.84EE331 pKa = 4.77EE332 pKa = 3.9YY333 pKa = 10.6HH334 pKa = 7.44FDD336 pKa = 3.55FKK338 pKa = 10.91MLNDD342 pKa = 4.02GDD344 pKa = 4.67DD345 pKa = 3.77LVIVCDD351 pKa = 3.56RR352 pKa = 11.84VTAKK356 pKa = 10.53AIKK359 pKa = 10.18DD360 pKa = 3.5KK361 pKa = 11.23LPSFFLKK368 pKa = 10.67FGIEE372 pKa = 3.98MEE374 pKa = 4.34FEE376 pKa = 4.47GLFHH380 pKa = 6.38TLEE383 pKa = 5.22AIEE386 pKa = 4.71FCQAHH391 pKa = 6.21PVQYY395 pKa = 10.91DD396 pKa = 3.25VGKK399 pKa = 9.71WIMVPNPSKK408 pKa = 10.93RR409 pKa = 11.84IFSDD413 pKa = 3.64LLSEE417 pKa = 4.65KK418 pKa = 9.98PVEE421 pKa = 4.06SRR423 pKa = 11.84KK424 pKa = 9.88IYY426 pKa = 10.41SSWLGAVGKK435 pKa = 9.23CGCAMSPGVPILQEE449 pKa = 4.06FYY451 pKa = 11.23DD452 pKa = 4.22WILRR456 pKa = 11.84TPGVKK461 pKa = 9.54PWIPKK466 pKa = 9.18EE467 pKa = 3.34GSYY470 pKa = 10.34YY471 pKa = 10.55YY472 pKa = 10.61KK473 pKa = 9.98YY474 pKa = 10.74GYY476 pKa = 10.11RR477 pKa = 11.84KK478 pKa = 9.88VIEE481 pKa = 4.14GAKK484 pKa = 9.08YY485 pKa = 10.33KK486 pKa = 10.12PVSEE490 pKa = 4.15IARR493 pKa = 11.84ISFFKK498 pKa = 10.5AYY500 pKa = 9.96NIPPSEE506 pKa = 4.02QIMLEE511 pKa = 3.85NMFRR515 pKa = 11.84SLPSPSRR522 pKa = 11.84EE523 pKa = 3.6ILEE526 pKa = 4.26EE527 pKa = 3.94SSRR530 pKa = 11.84PAHH533 pKa = 5.55TLPLEE538 pKa = 4.23MFALPTYY545 pKa = 9.97FKK547 pKa = 10.71KK548 pKa = 10.98FNWEE552 pKa = 3.38EE553 pKa = 3.74TDD555 pKa = 4.36RR556 pKa = 11.84MNRR559 pKa = 11.84WEE561 pKa = 4.28LDD563 pKa = 2.93RR564 pKa = 11.84GLFLVKK570 pKa = 10.26NKK572 pKa = 9.51RR573 pKa = 11.84RR574 pKa = 11.84AQDD577 pKa = 3.06KK578 pKa = 10.42GKK580 pKa = 11.32SNLTVKK586 pKa = 10.1NVKK589 pKa = 9.72KK590 pKa = 10.75DD591 pKa = 3.85PIRR594 pKa = 11.84FRR596 pKa = 11.84KK597 pKa = 9.02PKK599 pKa = 9.08RR600 pKa = 11.84SQRR603 pKa = 11.84RR604 pKa = 11.84RR605 pKa = 11.84ARR607 pKa = 11.84YY608 pKa = 6.61TSKK611 pKa = 10.35HH612 pKa = 5.26CRR614 pKa = 11.84HH615 pKa = 6.48RR616 pKa = 11.84SAKK619 pKa = 8.8TRR621 pKa = 11.84SAVTNLTKK629 pKa = 10.4SSRR632 pKa = 11.84QKK634 pKa = 10.32RR635 pKa = 11.84RR636 pKa = 11.84RR637 pKa = 11.84SAYY640 pKa = 8.48RR641 pKa = 11.84FRR643 pKa = 11.84YY644 pKa = 9.59RR645 pKa = 11.84SAA647 pKa = 3.46
Molecular weight: 75.57 kDa
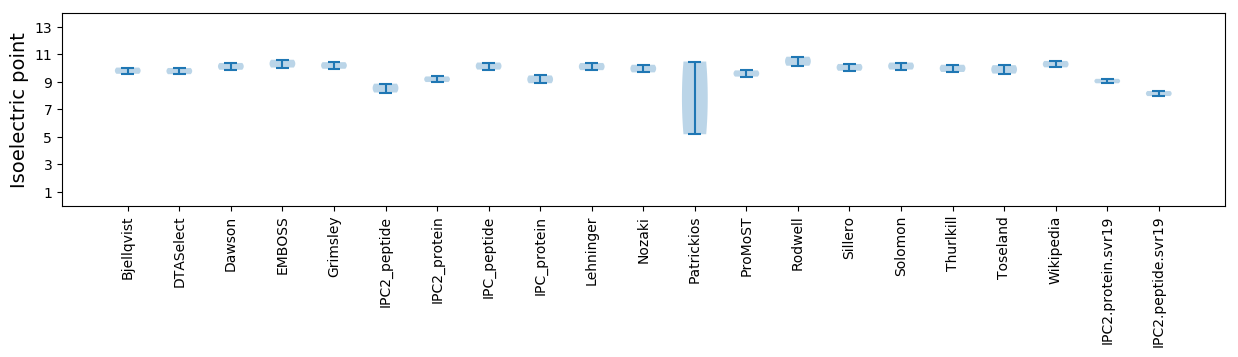
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFQ7|A0A1L3KFQ7_9VIRU Uncharacterized protein OS=Beihai tombus-like virus 2 OX=1922723 PE=4 SV=1
MM1 pKa = 7.47EE2 pKa = 4.35KK3 pKa = 10.38VIRR6 pKa = 11.84KK7 pKa = 4.91VTHH10 pKa = 5.93VIVSQRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84CAKK21 pKa = 10.33VIAQFTGRR29 pKa = 11.84CFRR32 pKa = 11.84CLAYY36 pKa = 10.38RR37 pKa = 11.84ATQLTQCRR45 pKa = 11.84VKK47 pKa = 10.43VIRR50 pKa = 11.84GAVVGTLKK58 pKa = 10.43RR59 pKa = 11.84VKK61 pKa = 9.76GQSRR65 pKa = 11.84RR66 pKa = 11.84LLVYY70 pKa = 10.32KK71 pKa = 10.32IRR73 pKa = 11.84NACRR77 pKa = 11.84GDD79 pKa = 3.32LKK81 pKa = 11.21LRR83 pKa = 11.84FQGTYY88 pKa = 9.92KK89 pKa = 10.54AVEE92 pKa = 4.5TIVDD96 pKa = 3.86RR97 pKa = 11.84TGHH100 pKa = 6.58AIEE103 pKa = 5.92IILGWMLQIFLAWFALFLMYY123 pKa = 10.88KK124 pKa = 10.33LGVLMNHH131 pKa = 7.11IYY133 pKa = 10.9LNYY136 pKa = 9.72IKK138 pKa = 10.37PEE140 pKa = 4.09YY141 pKa = 10.17EE142 pKa = 4.59SIATDD147 pKa = 3.15IPTKK151 pKa = 9.56MKK153 pKa = 9.76EE154 pKa = 3.97RR155 pKa = 11.84QRR157 pKa = 11.84LVYY160 pKa = 10.52CGMTWRR166 pKa = 11.84RR167 pKa = 11.84KK168 pKa = 8.62LMHH171 pKa = 6.51WRR173 pKa = 11.84MQLARR178 pKa = 11.84FFKK181 pKa = 10.53RR182 pKa = 11.84PVSGVEE188 pKa = 3.81PTPVTEE194 pKa = 4.13QIKK197 pKa = 10.22VLEE200 pKa = 4.38ALFEE204 pKa = 4.93ADD206 pKa = 4.18QLHH209 pKa = 6.56EE210 pKa = 4.93LPPLCTFRR218 pKa = 11.84GKK220 pKa = 9.96IVFSYY225 pKa = 10.96HH226 pKa = 6.43LAFKK230 pKa = 10.12MKK232 pKa = 9.49ATLGVVGKK240 pKa = 10.78YY241 pKa = 10.0DD242 pKa = 3.42VARR245 pKa = 11.84TNVLIRR251 pKa = 11.84TANAILVEE259 pKa = 4.23MLQEE263 pKa = 3.96MSDD266 pKa = 3.72EE267 pKa = 4.07EE268 pKa = 4.4KK269 pKa = 11.05EE270 pKa = 4.07RR271 pKa = 11.84VKK273 pKa = 10.74LLKK276 pKa = 10.28YY277 pKa = 10.45NHH279 pKa = 6.19GLRR282 pKa = 11.84HH283 pKa = 5.71KK284 pKa = 9.5VVCQAIAIFKK294 pKa = 10.17IPSFDD299 pKa = 3.71EE300 pKa = 3.84VSMAEE305 pKa = 4.14DD306 pKa = 3.34LRR308 pKa = 11.84ASVLSQITDD317 pKa = 3.24AFVSKK322 pKa = 11.0VGGPSNN328 pKa = 3.55
MM1 pKa = 7.47EE2 pKa = 4.35KK3 pKa = 10.38VIRR6 pKa = 11.84KK7 pKa = 4.91VTHH10 pKa = 5.93VIVSQRR16 pKa = 11.84RR17 pKa = 11.84RR18 pKa = 11.84CAKK21 pKa = 10.33VIAQFTGRR29 pKa = 11.84CFRR32 pKa = 11.84CLAYY36 pKa = 10.38RR37 pKa = 11.84ATQLTQCRR45 pKa = 11.84VKK47 pKa = 10.43VIRR50 pKa = 11.84GAVVGTLKK58 pKa = 10.43RR59 pKa = 11.84VKK61 pKa = 9.76GQSRR65 pKa = 11.84RR66 pKa = 11.84LLVYY70 pKa = 10.32KK71 pKa = 10.32IRR73 pKa = 11.84NACRR77 pKa = 11.84GDD79 pKa = 3.32LKK81 pKa = 11.21LRR83 pKa = 11.84FQGTYY88 pKa = 9.92KK89 pKa = 10.54AVEE92 pKa = 4.5TIVDD96 pKa = 3.86RR97 pKa = 11.84TGHH100 pKa = 6.58AIEE103 pKa = 5.92IILGWMLQIFLAWFALFLMYY123 pKa = 10.88KK124 pKa = 10.33LGVLMNHH131 pKa = 7.11IYY133 pKa = 10.9LNYY136 pKa = 9.72IKK138 pKa = 10.37PEE140 pKa = 4.09YY141 pKa = 10.17EE142 pKa = 4.59SIATDD147 pKa = 3.15IPTKK151 pKa = 9.56MKK153 pKa = 9.76EE154 pKa = 3.97RR155 pKa = 11.84QRR157 pKa = 11.84LVYY160 pKa = 10.52CGMTWRR166 pKa = 11.84RR167 pKa = 11.84KK168 pKa = 8.62LMHH171 pKa = 6.51WRR173 pKa = 11.84MQLARR178 pKa = 11.84FFKK181 pKa = 10.53RR182 pKa = 11.84PVSGVEE188 pKa = 3.81PTPVTEE194 pKa = 4.13QIKK197 pKa = 10.22VLEE200 pKa = 4.38ALFEE204 pKa = 4.93ADD206 pKa = 4.18QLHH209 pKa = 6.56EE210 pKa = 4.93LPPLCTFRR218 pKa = 11.84GKK220 pKa = 9.96IVFSYY225 pKa = 10.96HH226 pKa = 6.43LAFKK230 pKa = 10.12MKK232 pKa = 9.49ATLGVVGKK240 pKa = 10.78YY241 pKa = 10.0DD242 pKa = 3.42VARR245 pKa = 11.84TNVLIRR251 pKa = 11.84TANAILVEE259 pKa = 4.23MLQEE263 pKa = 3.96MSDD266 pKa = 3.72EE267 pKa = 4.07EE268 pKa = 4.4KK269 pKa = 11.05EE270 pKa = 4.07RR271 pKa = 11.84VKK273 pKa = 10.74LLKK276 pKa = 10.28YY277 pKa = 10.45NHH279 pKa = 6.19GLRR282 pKa = 11.84HH283 pKa = 5.71KK284 pKa = 9.5VVCQAIAIFKK294 pKa = 10.17IPSFDD299 pKa = 3.71EE300 pKa = 3.84VSMAEE305 pKa = 4.14DD306 pKa = 3.34LRR308 pKa = 11.84ASVLSQITDD317 pKa = 3.24AFVSKK322 pKa = 11.0VGGPSNN328 pKa = 3.55
Molecular weight: 37.89 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
975 |
328 |
647 |
487.5 |
56.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.949 ± 0.761 | 1.846 ± 0.33 |
4.513 ± 0.984 | 5.949 ± 0.256 |
4.718 ± 0.081 | 4.821 ± 0.202 |
2.359 ± 0.045 | 6.564 ± 0.08 |
8.0 ± 0.041 | 8.205 ± 1.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.282 ± 0.21 | 3.179 ± 0.582 |
4.615 ± 1.041 | 3.59 ± 0.378 |
8.615 ± 0.126 | 5.744 ± 1.16 |
4.821 ± 0.371 | 7.59 ± 1.206 |
1.538 ± 0.177 | 4.103 ± 0.417 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
