
Botrytis ourmia-like virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Lenarviricota; Miaviricetes; Ourlivirales; Botourmiaviridae; Botoulivirus; Botrytis botoulivirus
Average proteome isoelectric point is 8.68
Get precalculated fractions of proteins
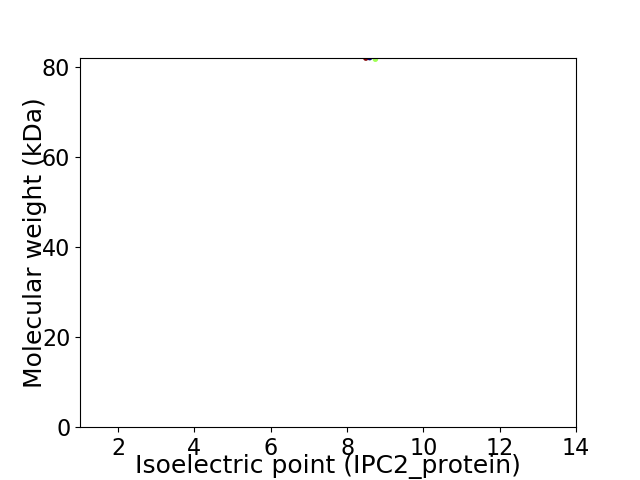
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
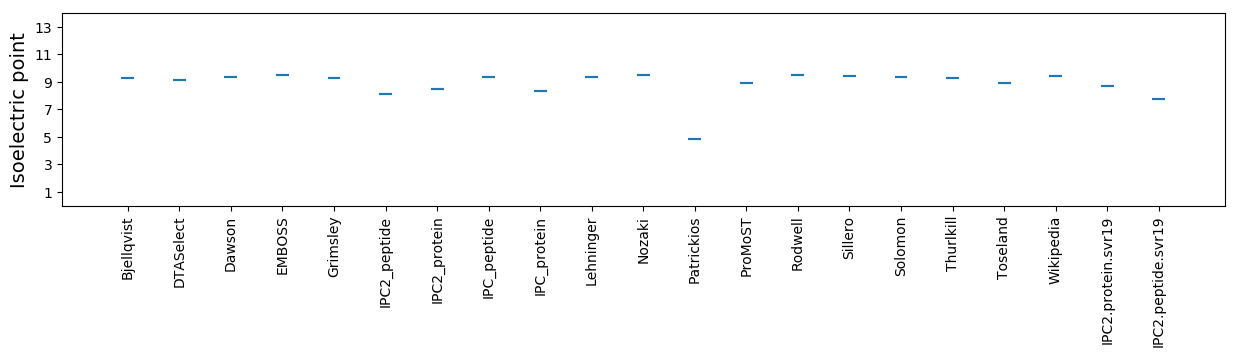
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
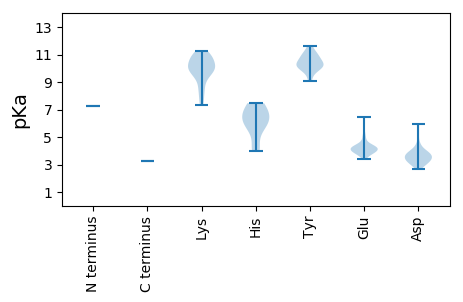
Protein with the lowest isoelectric point:
>tr|A0A0S4GA80|A0A0S4GA80_9VIRU RNA dependent RNA polymerase OS=Botrytis ourmia-like virus OX=1872719 GN=RdRp PE=4 SV=1
MM1 pKa = 7.22PTSNDD6 pKa = 2.97MEE8 pKa = 4.97AGCKK12 pKa = 7.73ATRR15 pKa = 11.84SIRR18 pKa = 11.84EE19 pKa = 3.87SLLRR23 pKa = 11.84ATQLISRR30 pKa = 11.84EE31 pKa = 4.01FNLPVPVGPFNGTSCMEE48 pKa = 5.05LRR50 pKa = 11.84SQWDD54 pKa = 3.95TYY56 pKa = 11.65AKK58 pKa = 9.61WSCLQFVSKK67 pKa = 10.7KK68 pKa = 7.33EE69 pKa = 3.96EE70 pKa = 3.8RR71 pKa = 11.84RR72 pKa = 11.84KK73 pKa = 8.43TLVQLIKK80 pKa = 10.56SGCRR84 pKa = 11.84LFDD87 pKa = 4.07GDD89 pKa = 5.98CRR91 pKa = 11.84TCDD94 pKa = 3.43PKK96 pKa = 11.16LKK98 pKa = 10.23EE99 pKa = 3.84LEE101 pKa = 3.83KK102 pKa = 10.76RR103 pKa = 11.84KK104 pKa = 9.19WVEE107 pKa = 3.61QMSTARR113 pKa = 11.84PRR115 pKa = 11.84VAYY118 pKa = 10.27YY119 pKa = 10.52KK120 pKa = 10.87GLTRR124 pKa = 11.84STWLGRR130 pKa = 11.84LKK132 pKa = 10.9DD133 pKa = 3.65SARR136 pKa = 11.84EE137 pKa = 4.01LITGWGRR144 pKa = 11.84NLEE147 pKa = 4.18GCRR150 pKa = 11.84MEE152 pKa = 6.43DD153 pKa = 3.93YY154 pKa = 10.97IPDD157 pKa = 3.36QSGCFEE163 pKa = 4.37AKK165 pKa = 10.22SIEE168 pKa = 4.21GGTLSVSQDD177 pKa = 3.09FASFPNEE184 pKa = 3.73VRR186 pKa = 11.84LGTAKK191 pKa = 9.86TKK193 pKa = 10.66GKK195 pKa = 9.98IRR197 pKa = 11.84VVTMQGANTKK207 pKa = 9.78KK208 pKa = 9.96VLRR211 pKa = 11.84PVHH214 pKa = 5.9SALYY218 pKa = 10.14DD219 pKa = 3.62YY220 pKa = 11.21LAGFGWLVRR229 pKa = 11.84GDD231 pKa = 3.23VTAADD236 pKa = 4.44FEE238 pKa = 5.4AIIEE242 pKa = 4.31DD243 pKa = 3.65KK244 pKa = 11.22QGDD247 pKa = 3.83EE248 pKa = 4.45KK249 pKa = 11.05FISGDD254 pKa = 3.35YY255 pKa = 9.93EE256 pKa = 3.84QATNHH261 pKa = 5.77INIDD265 pKa = 3.63SVQAIISVIAEE276 pKa = 4.08EE277 pKa = 4.48PLLSDD282 pKa = 3.84EE283 pKa = 4.26EE284 pKa = 4.79RR285 pKa = 11.84EE286 pKa = 3.97VLIRR290 pKa = 11.84SFRR293 pKa = 11.84DD294 pKa = 3.05VTVYY298 pKa = 10.96KK299 pKa = 11.04NNGSFLCKK307 pKa = 10.03VRR309 pKa = 11.84NGSMMGNLVSFPLLCILNKK328 pKa = 10.09CCYY331 pKa = 10.32DD332 pKa = 3.23MSRR335 pKa = 11.84EE336 pKa = 4.0IEE338 pKa = 4.14SEE340 pKa = 4.26EE341 pKa = 4.26NGVPYY346 pKa = 10.2CPRR349 pKa = 11.84VGRR352 pKa = 11.84FNGDD356 pKa = 3.04DD357 pKa = 3.63CAFCGTDD364 pKa = 2.68RR365 pKa = 11.84FFEE368 pKa = 4.06IWRR371 pKa = 11.84EE372 pKa = 3.94TTSIFGLVVQEE383 pKa = 4.29KK384 pKa = 8.57KK385 pKa = 9.37TGISSRR391 pKa = 11.84WIEE394 pKa = 4.03LNSEE398 pKa = 4.16SFDD401 pKa = 3.79SLKK404 pKa = 10.83HH405 pKa = 6.35RR406 pKa = 11.84FVQKK410 pKa = 10.94NFLSYY415 pKa = 11.03LRR417 pKa = 11.84VTRR420 pKa = 11.84DD421 pKa = 3.15TPGDD425 pKa = 3.73LLSEE429 pKa = 4.22IVSGTKK435 pKa = 10.11GFKK438 pKa = 10.61GSTKK442 pKa = 8.54MWLINHH448 pKa = 5.8VLRR451 pKa = 11.84YY452 pKa = 9.07EE453 pKa = 3.72IAIRR457 pKa = 11.84GVCASTIPRR466 pKa = 11.84KK467 pKa = 10.18LFLLLIKK474 pKa = 10.27RR475 pKa = 11.84VWFRR479 pKa = 11.84KK480 pKa = 7.56TLSNPLPPFPTTGVDD495 pKa = 2.98RR496 pKa = 11.84SIRR499 pKa = 11.84QTVVSPPLPSFIPLIDD515 pKa = 5.29DD516 pKa = 3.5IEE518 pKa = 4.4TLVRR522 pKa = 11.84KK523 pKa = 9.48SHH525 pKa = 4.27VKK527 pKa = 9.42KK528 pKa = 9.6WVGVSTIDD536 pKa = 3.87GRR538 pKa = 11.84RR539 pKa = 11.84ARR541 pKa = 11.84CSNEE545 pKa = 3.42MNPDD549 pKa = 3.32HH550 pKa = 7.46DD551 pKa = 3.98HH552 pKa = 6.28FRR554 pKa = 11.84PMVHH558 pKa = 6.65GSLFFPFISSSYY570 pKa = 10.71SPTPLSSSLRR580 pKa = 11.84GSLLRR585 pKa = 11.84QRR587 pKa = 11.84KK588 pKa = 8.19VAFDD592 pKa = 4.49LSSKK596 pKa = 9.99PSASVRR602 pKa = 11.84FSRR605 pKa = 11.84KK606 pKa = 9.11HH607 pKa = 3.98SWGFTMSEE615 pKa = 4.2LCKK618 pKa = 10.35DD619 pKa = 3.33TLTTFFGEE627 pKa = 4.3TWLMPPKK634 pKa = 9.91SLCGSTLPYY643 pKa = 9.57YY644 pKa = 10.46HH645 pKa = 7.38PHH647 pKa = 6.73LKK649 pKa = 9.98IKK651 pKa = 10.52CDD653 pKa = 3.66FLTTVTPTYY662 pKa = 10.04YY663 pKa = 9.97PPPVSLLTTKK673 pKa = 9.91TLAEE677 pKa = 4.2HH678 pKa = 7.0AATIYY683 pKa = 10.58NLEE686 pKa = 4.24RR687 pKa = 11.84TSSSILRR694 pKa = 11.84NSRR697 pKa = 11.84VRR699 pKa = 11.84IAKK702 pKa = 9.81ADD704 pKa = 3.52SKK706 pKa = 10.91LWSFVPDD713 pKa = 3.41NYY715 pKa = 10.1IGSRR719 pKa = 11.84LVVV722 pKa = 3.28
MM1 pKa = 7.22PTSNDD6 pKa = 2.97MEE8 pKa = 4.97AGCKK12 pKa = 7.73ATRR15 pKa = 11.84SIRR18 pKa = 11.84EE19 pKa = 3.87SLLRR23 pKa = 11.84ATQLISRR30 pKa = 11.84EE31 pKa = 4.01FNLPVPVGPFNGTSCMEE48 pKa = 5.05LRR50 pKa = 11.84SQWDD54 pKa = 3.95TYY56 pKa = 11.65AKK58 pKa = 9.61WSCLQFVSKK67 pKa = 10.7KK68 pKa = 7.33EE69 pKa = 3.96EE70 pKa = 3.8RR71 pKa = 11.84RR72 pKa = 11.84KK73 pKa = 8.43TLVQLIKK80 pKa = 10.56SGCRR84 pKa = 11.84LFDD87 pKa = 4.07GDD89 pKa = 5.98CRR91 pKa = 11.84TCDD94 pKa = 3.43PKK96 pKa = 11.16LKK98 pKa = 10.23EE99 pKa = 3.84LEE101 pKa = 3.83KK102 pKa = 10.76RR103 pKa = 11.84KK104 pKa = 9.19WVEE107 pKa = 3.61QMSTARR113 pKa = 11.84PRR115 pKa = 11.84VAYY118 pKa = 10.27YY119 pKa = 10.52KK120 pKa = 10.87GLTRR124 pKa = 11.84STWLGRR130 pKa = 11.84LKK132 pKa = 10.9DD133 pKa = 3.65SARR136 pKa = 11.84EE137 pKa = 4.01LITGWGRR144 pKa = 11.84NLEE147 pKa = 4.18GCRR150 pKa = 11.84MEE152 pKa = 6.43DD153 pKa = 3.93YY154 pKa = 10.97IPDD157 pKa = 3.36QSGCFEE163 pKa = 4.37AKK165 pKa = 10.22SIEE168 pKa = 4.21GGTLSVSQDD177 pKa = 3.09FASFPNEE184 pKa = 3.73VRR186 pKa = 11.84LGTAKK191 pKa = 9.86TKK193 pKa = 10.66GKK195 pKa = 9.98IRR197 pKa = 11.84VVTMQGANTKK207 pKa = 9.78KK208 pKa = 9.96VLRR211 pKa = 11.84PVHH214 pKa = 5.9SALYY218 pKa = 10.14DD219 pKa = 3.62YY220 pKa = 11.21LAGFGWLVRR229 pKa = 11.84GDD231 pKa = 3.23VTAADD236 pKa = 4.44FEE238 pKa = 5.4AIIEE242 pKa = 4.31DD243 pKa = 3.65KK244 pKa = 11.22QGDD247 pKa = 3.83EE248 pKa = 4.45KK249 pKa = 11.05FISGDD254 pKa = 3.35YY255 pKa = 9.93EE256 pKa = 3.84QATNHH261 pKa = 5.77INIDD265 pKa = 3.63SVQAIISVIAEE276 pKa = 4.08EE277 pKa = 4.48PLLSDD282 pKa = 3.84EE283 pKa = 4.26EE284 pKa = 4.79RR285 pKa = 11.84EE286 pKa = 3.97VLIRR290 pKa = 11.84SFRR293 pKa = 11.84DD294 pKa = 3.05VTVYY298 pKa = 10.96KK299 pKa = 11.04NNGSFLCKK307 pKa = 10.03VRR309 pKa = 11.84NGSMMGNLVSFPLLCILNKK328 pKa = 10.09CCYY331 pKa = 10.32DD332 pKa = 3.23MSRR335 pKa = 11.84EE336 pKa = 4.0IEE338 pKa = 4.14SEE340 pKa = 4.26EE341 pKa = 4.26NGVPYY346 pKa = 10.2CPRR349 pKa = 11.84VGRR352 pKa = 11.84FNGDD356 pKa = 3.04DD357 pKa = 3.63CAFCGTDD364 pKa = 2.68RR365 pKa = 11.84FFEE368 pKa = 4.06IWRR371 pKa = 11.84EE372 pKa = 3.94TTSIFGLVVQEE383 pKa = 4.29KK384 pKa = 8.57KK385 pKa = 9.37TGISSRR391 pKa = 11.84WIEE394 pKa = 4.03LNSEE398 pKa = 4.16SFDD401 pKa = 3.79SLKK404 pKa = 10.83HH405 pKa = 6.35RR406 pKa = 11.84FVQKK410 pKa = 10.94NFLSYY415 pKa = 11.03LRR417 pKa = 11.84VTRR420 pKa = 11.84DD421 pKa = 3.15TPGDD425 pKa = 3.73LLSEE429 pKa = 4.22IVSGTKK435 pKa = 10.11GFKK438 pKa = 10.61GSTKK442 pKa = 8.54MWLINHH448 pKa = 5.8VLRR451 pKa = 11.84YY452 pKa = 9.07EE453 pKa = 3.72IAIRR457 pKa = 11.84GVCASTIPRR466 pKa = 11.84KK467 pKa = 10.18LFLLLIKK474 pKa = 10.27RR475 pKa = 11.84VWFRR479 pKa = 11.84KK480 pKa = 7.56TLSNPLPPFPTTGVDD495 pKa = 2.98RR496 pKa = 11.84SIRR499 pKa = 11.84QTVVSPPLPSFIPLIDD515 pKa = 5.29DD516 pKa = 3.5IEE518 pKa = 4.4TLVRR522 pKa = 11.84KK523 pKa = 9.48SHH525 pKa = 4.27VKK527 pKa = 9.42KK528 pKa = 9.6WVGVSTIDD536 pKa = 3.87GRR538 pKa = 11.84RR539 pKa = 11.84ARR541 pKa = 11.84CSNEE545 pKa = 3.42MNPDD549 pKa = 3.32HH550 pKa = 7.46DD551 pKa = 3.98HH552 pKa = 6.28FRR554 pKa = 11.84PMVHH558 pKa = 6.65GSLFFPFISSSYY570 pKa = 10.71SPTPLSSSLRR580 pKa = 11.84GSLLRR585 pKa = 11.84QRR587 pKa = 11.84KK588 pKa = 8.19VAFDD592 pKa = 4.49LSSKK596 pKa = 9.99PSASVRR602 pKa = 11.84FSRR605 pKa = 11.84KK606 pKa = 9.11HH607 pKa = 3.98SWGFTMSEE615 pKa = 4.2LCKK618 pKa = 10.35DD619 pKa = 3.33TLTTFFGEE627 pKa = 4.3TWLMPPKK634 pKa = 9.91SLCGSTLPYY643 pKa = 9.57YY644 pKa = 10.46HH645 pKa = 7.38PHH647 pKa = 6.73LKK649 pKa = 9.98IKK651 pKa = 10.52CDD653 pKa = 3.66FLTTVTPTYY662 pKa = 10.04YY663 pKa = 9.97PPPVSLLTTKK673 pKa = 9.91TLAEE677 pKa = 4.2HH678 pKa = 7.0AATIYY683 pKa = 10.58NLEE686 pKa = 4.24RR687 pKa = 11.84TSSSILRR694 pKa = 11.84NSRR697 pKa = 11.84VRR699 pKa = 11.84IAKK702 pKa = 9.81ADD704 pKa = 3.52SKK706 pKa = 10.91LWSFVPDD713 pKa = 3.41NYY715 pKa = 10.1IGSRR719 pKa = 11.84LVVV722 pKa = 3.28
Molecular weight: 81.88 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0S4GA80|A0A0S4GA80_9VIRU RNA dependent RNA polymerase OS=Botrytis ourmia-like virus OX=1872719 GN=RdRp PE=4 SV=1
MM1 pKa = 7.22PTSNDD6 pKa = 2.97MEE8 pKa = 4.97AGCKK12 pKa = 7.73ATRR15 pKa = 11.84SIRR18 pKa = 11.84EE19 pKa = 3.87SLLRR23 pKa = 11.84ATQLISRR30 pKa = 11.84EE31 pKa = 4.01FNLPVPVGPFNGTSCMEE48 pKa = 5.05LRR50 pKa = 11.84SQWDD54 pKa = 3.95TYY56 pKa = 11.65AKK58 pKa = 9.61WSCLQFVSKK67 pKa = 10.7KK68 pKa = 7.33EE69 pKa = 3.96EE70 pKa = 3.8RR71 pKa = 11.84RR72 pKa = 11.84KK73 pKa = 8.43TLVQLIKK80 pKa = 10.56SGCRR84 pKa = 11.84LFDD87 pKa = 4.07GDD89 pKa = 5.98CRR91 pKa = 11.84TCDD94 pKa = 3.43PKK96 pKa = 11.16LKK98 pKa = 10.23EE99 pKa = 3.84LEE101 pKa = 3.83KK102 pKa = 10.76RR103 pKa = 11.84KK104 pKa = 9.19WVEE107 pKa = 3.61QMSTARR113 pKa = 11.84PRR115 pKa = 11.84VAYY118 pKa = 10.27YY119 pKa = 10.52KK120 pKa = 10.87GLTRR124 pKa = 11.84STWLGRR130 pKa = 11.84LKK132 pKa = 10.9DD133 pKa = 3.65SARR136 pKa = 11.84EE137 pKa = 4.01LITGWGRR144 pKa = 11.84NLEE147 pKa = 4.18GCRR150 pKa = 11.84MEE152 pKa = 6.43DD153 pKa = 3.93YY154 pKa = 10.97IPDD157 pKa = 3.36QSGCFEE163 pKa = 4.37AKK165 pKa = 10.22SIEE168 pKa = 4.21GGTLSVSQDD177 pKa = 3.09FASFPNEE184 pKa = 3.73VRR186 pKa = 11.84LGTAKK191 pKa = 9.86TKK193 pKa = 10.66GKK195 pKa = 9.98IRR197 pKa = 11.84VVTMQGANTKK207 pKa = 9.78KK208 pKa = 9.96VLRR211 pKa = 11.84PVHH214 pKa = 5.9SALYY218 pKa = 10.14DD219 pKa = 3.62YY220 pKa = 11.21LAGFGWLVRR229 pKa = 11.84GDD231 pKa = 3.23VTAADD236 pKa = 4.44FEE238 pKa = 5.4AIIEE242 pKa = 4.31DD243 pKa = 3.65KK244 pKa = 11.22QGDD247 pKa = 3.83EE248 pKa = 4.45KK249 pKa = 11.05FISGDD254 pKa = 3.35YY255 pKa = 9.93EE256 pKa = 3.84QATNHH261 pKa = 5.77INIDD265 pKa = 3.63SVQAIISVIAEE276 pKa = 4.08EE277 pKa = 4.48PLLSDD282 pKa = 3.84EE283 pKa = 4.26EE284 pKa = 4.79RR285 pKa = 11.84EE286 pKa = 3.97VLIRR290 pKa = 11.84SFRR293 pKa = 11.84DD294 pKa = 3.05VTVYY298 pKa = 10.96KK299 pKa = 11.04NNGSFLCKK307 pKa = 10.03VRR309 pKa = 11.84NGSMMGNLVSFPLLCILNKK328 pKa = 10.09CCYY331 pKa = 10.32DD332 pKa = 3.23MSRR335 pKa = 11.84EE336 pKa = 4.0IEE338 pKa = 4.14SEE340 pKa = 4.26EE341 pKa = 4.26NGVPYY346 pKa = 10.2CPRR349 pKa = 11.84VGRR352 pKa = 11.84FNGDD356 pKa = 3.04DD357 pKa = 3.63CAFCGTDD364 pKa = 2.68RR365 pKa = 11.84FFEE368 pKa = 4.06IWRR371 pKa = 11.84EE372 pKa = 3.94TTSIFGLVVQEE383 pKa = 4.29KK384 pKa = 8.57KK385 pKa = 9.37TGISSRR391 pKa = 11.84WIEE394 pKa = 4.03LNSEE398 pKa = 4.16SFDD401 pKa = 3.79SLKK404 pKa = 10.83HH405 pKa = 6.35RR406 pKa = 11.84FVQKK410 pKa = 10.94NFLSYY415 pKa = 11.03LRR417 pKa = 11.84VTRR420 pKa = 11.84DD421 pKa = 3.15TPGDD425 pKa = 3.73LLSEE429 pKa = 4.22IVSGTKK435 pKa = 10.11GFKK438 pKa = 10.61GSTKK442 pKa = 8.54MWLINHH448 pKa = 5.8VLRR451 pKa = 11.84YY452 pKa = 9.07EE453 pKa = 3.72IAIRR457 pKa = 11.84GVCASTIPRR466 pKa = 11.84KK467 pKa = 10.18LFLLLIKK474 pKa = 10.27RR475 pKa = 11.84VWFRR479 pKa = 11.84KK480 pKa = 7.56TLSNPLPPFPTTGVDD495 pKa = 2.98RR496 pKa = 11.84SIRR499 pKa = 11.84QTVVSPPLPSFIPLIDD515 pKa = 5.29DD516 pKa = 3.5IEE518 pKa = 4.4TLVRR522 pKa = 11.84KK523 pKa = 9.48SHH525 pKa = 4.27VKK527 pKa = 9.42KK528 pKa = 9.6WVGVSTIDD536 pKa = 3.87GRR538 pKa = 11.84RR539 pKa = 11.84ARR541 pKa = 11.84CSNEE545 pKa = 3.42MNPDD549 pKa = 3.32HH550 pKa = 7.46DD551 pKa = 3.98HH552 pKa = 6.28FRR554 pKa = 11.84PMVHH558 pKa = 6.65GSLFFPFISSSYY570 pKa = 10.71SPTPLSSSLRR580 pKa = 11.84GSLLRR585 pKa = 11.84QRR587 pKa = 11.84KK588 pKa = 8.19VAFDD592 pKa = 4.49LSSKK596 pKa = 9.99PSASVRR602 pKa = 11.84FSRR605 pKa = 11.84KK606 pKa = 9.11HH607 pKa = 3.98SWGFTMSEE615 pKa = 4.2LCKK618 pKa = 10.35DD619 pKa = 3.33TLTTFFGEE627 pKa = 4.3TWLMPPKK634 pKa = 9.91SLCGSTLPYY643 pKa = 9.57YY644 pKa = 10.46HH645 pKa = 7.38PHH647 pKa = 6.73LKK649 pKa = 9.98IKK651 pKa = 10.52CDD653 pKa = 3.66FLTTVTPTYY662 pKa = 10.04YY663 pKa = 9.97PPPVSLLTTKK673 pKa = 9.91TLAEE677 pKa = 4.2HH678 pKa = 7.0AATIYY683 pKa = 10.58NLEE686 pKa = 4.24RR687 pKa = 11.84TSSSILRR694 pKa = 11.84NSRR697 pKa = 11.84VRR699 pKa = 11.84IAKK702 pKa = 9.81ADD704 pKa = 3.52SKK706 pKa = 10.91LWSFVPDD713 pKa = 3.41NYY715 pKa = 10.1IGSRR719 pKa = 11.84LVVV722 pKa = 3.28
MM1 pKa = 7.22PTSNDD6 pKa = 2.97MEE8 pKa = 4.97AGCKK12 pKa = 7.73ATRR15 pKa = 11.84SIRR18 pKa = 11.84EE19 pKa = 3.87SLLRR23 pKa = 11.84ATQLISRR30 pKa = 11.84EE31 pKa = 4.01FNLPVPVGPFNGTSCMEE48 pKa = 5.05LRR50 pKa = 11.84SQWDD54 pKa = 3.95TYY56 pKa = 11.65AKK58 pKa = 9.61WSCLQFVSKK67 pKa = 10.7KK68 pKa = 7.33EE69 pKa = 3.96EE70 pKa = 3.8RR71 pKa = 11.84RR72 pKa = 11.84KK73 pKa = 8.43TLVQLIKK80 pKa = 10.56SGCRR84 pKa = 11.84LFDD87 pKa = 4.07GDD89 pKa = 5.98CRR91 pKa = 11.84TCDD94 pKa = 3.43PKK96 pKa = 11.16LKK98 pKa = 10.23EE99 pKa = 3.84LEE101 pKa = 3.83KK102 pKa = 10.76RR103 pKa = 11.84KK104 pKa = 9.19WVEE107 pKa = 3.61QMSTARR113 pKa = 11.84PRR115 pKa = 11.84VAYY118 pKa = 10.27YY119 pKa = 10.52KK120 pKa = 10.87GLTRR124 pKa = 11.84STWLGRR130 pKa = 11.84LKK132 pKa = 10.9DD133 pKa = 3.65SARR136 pKa = 11.84EE137 pKa = 4.01LITGWGRR144 pKa = 11.84NLEE147 pKa = 4.18GCRR150 pKa = 11.84MEE152 pKa = 6.43DD153 pKa = 3.93YY154 pKa = 10.97IPDD157 pKa = 3.36QSGCFEE163 pKa = 4.37AKK165 pKa = 10.22SIEE168 pKa = 4.21GGTLSVSQDD177 pKa = 3.09FASFPNEE184 pKa = 3.73VRR186 pKa = 11.84LGTAKK191 pKa = 9.86TKK193 pKa = 10.66GKK195 pKa = 9.98IRR197 pKa = 11.84VVTMQGANTKK207 pKa = 9.78KK208 pKa = 9.96VLRR211 pKa = 11.84PVHH214 pKa = 5.9SALYY218 pKa = 10.14DD219 pKa = 3.62YY220 pKa = 11.21LAGFGWLVRR229 pKa = 11.84GDD231 pKa = 3.23VTAADD236 pKa = 4.44FEE238 pKa = 5.4AIIEE242 pKa = 4.31DD243 pKa = 3.65KK244 pKa = 11.22QGDD247 pKa = 3.83EE248 pKa = 4.45KK249 pKa = 11.05FISGDD254 pKa = 3.35YY255 pKa = 9.93EE256 pKa = 3.84QATNHH261 pKa = 5.77INIDD265 pKa = 3.63SVQAIISVIAEE276 pKa = 4.08EE277 pKa = 4.48PLLSDD282 pKa = 3.84EE283 pKa = 4.26EE284 pKa = 4.79RR285 pKa = 11.84EE286 pKa = 3.97VLIRR290 pKa = 11.84SFRR293 pKa = 11.84DD294 pKa = 3.05VTVYY298 pKa = 10.96KK299 pKa = 11.04NNGSFLCKK307 pKa = 10.03VRR309 pKa = 11.84NGSMMGNLVSFPLLCILNKK328 pKa = 10.09CCYY331 pKa = 10.32DD332 pKa = 3.23MSRR335 pKa = 11.84EE336 pKa = 4.0IEE338 pKa = 4.14SEE340 pKa = 4.26EE341 pKa = 4.26NGVPYY346 pKa = 10.2CPRR349 pKa = 11.84VGRR352 pKa = 11.84FNGDD356 pKa = 3.04DD357 pKa = 3.63CAFCGTDD364 pKa = 2.68RR365 pKa = 11.84FFEE368 pKa = 4.06IWRR371 pKa = 11.84EE372 pKa = 3.94TTSIFGLVVQEE383 pKa = 4.29KK384 pKa = 8.57KK385 pKa = 9.37TGISSRR391 pKa = 11.84WIEE394 pKa = 4.03LNSEE398 pKa = 4.16SFDD401 pKa = 3.79SLKK404 pKa = 10.83HH405 pKa = 6.35RR406 pKa = 11.84FVQKK410 pKa = 10.94NFLSYY415 pKa = 11.03LRR417 pKa = 11.84VTRR420 pKa = 11.84DD421 pKa = 3.15TPGDD425 pKa = 3.73LLSEE429 pKa = 4.22IVSGTKK435 pKa = 10.11GFKK438 pKa = 10.61GSTKK442 pKa = 8.54MWLINHH448 pKa = 5.8VLRR451 pKa = 11.84YY452 pKa = 9.07EE453 pKa = 3.72IAIRR457 pKa = 11.84GVCASTIPRR466 pKa = 11.84KK467 pKa = 10.18LFLLLIKK474 pKa = 10.27RR475 pKa = 11.84VWFRR479 pKa = 11.84KK480 pKa = 7.56TLSNPLPPFPTTGVDD495 pKa = 2.98RR496 pKa = 11.84SIRR499 pKa = 11.84QTVVSPPLPSFIPLIDD515 pKa = 5.29DD516 pKa = 3.5IEE518 pKa = 4.4TLVRR522 pKa = 11.84KK523 pKa = 9.48SHH525 pKa = 4.27VKK527 pKa = 9.42KK528 pKa = 9.6WVGVSTIDD536 pKa = 3.87GRR538 pKa = 11.84RR539 pKa = 11.84ARR541 pKa = 11.84CSNEE545 pKa = 3.42MNPDD549 pKa = 3.32HH550 pKa = 7.46DD551 pKa = 3.98HH552 pKa = 6.28FRR554 pKa = 11.84PMVHH558 pKa = 6.65GSLFFPFISSSYY570 pKa = 10.71SPTPLSSSLRR580 pKa = 11.84GSLLRR585 pKa = 11.84QRR587 pKa = 11.84KK588 pKa = 8.19VAFDD592 pKa = 4.49LSSKK596 pKa = 9.99PSASVRR602 pKa = 11.84FSRR605 pKa = 11.84KK606 pKa = 9.11HH607 pKa = 3.98SWGFTMSEE615 pKa = 4.2LCKK618 pKa = 10.35DD619 pKa = 3.33TLTTFFGEE627 pKa = 4.3TWLMPPKK634 pKa = 9.91SLCGSTLPYY643 pKa = 9.57YY644 pKa = 10.46HH645 pKa = 7.38PHH647 pKa = 6.73LKK649 pKa = 9.98IKK651 pKa = 10.52CDD653 pKa = 3.66FLTTVTPTYY662 pKa = 10.04YY663 pKa = 9.97PPPVSLLTTKK673 pKa = 9.91TLAEE677 pKa = 4.2HH678 pKa = 7.0AATIYY683 pKa = 10.58NLEE686 pKa = 4.24RR687 pKa = 11.84TSSSILRR694 pKa = 11.84NSRR697 pKa = 11.84VRR699 pKa = 11.84IAKK702 pKa = 9.81ADD704 pKa = 3.52SKK706 pKa = 10.91LWSFVPDD713 pKa = 3.41NYY715 pKa = 10.1IGSRR719 pKa = 11.84LVVV722 pKa = 3.28
Molecular weight: 81.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
722 |
722 |
722 |
722.0 |
81.88 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.155 ± 0.0 | 2.77 ± 0.0 |
4.986 ± 0.0 | 5.679 ± 0.0 |
5.125 ± 0.0 | 6.094 ± 0.0 |
1.662 ± 0.0 | 5.402 ± 0.0 |
6.371 ± 0.0 | 9.418 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.939 ± 0.0 | 3.324 ± 0.0 |
5.263 ± 0.0 | 2.078 ± 0.0 |
7.756 ± 0.0 | 9.972 ± 0.0 |
6.925 ± 0.0 | 6.51 ± 0.0 |
1.939 ± 0.0 | 2.632 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
