
Celeribacter neptunius
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Celeribacter
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
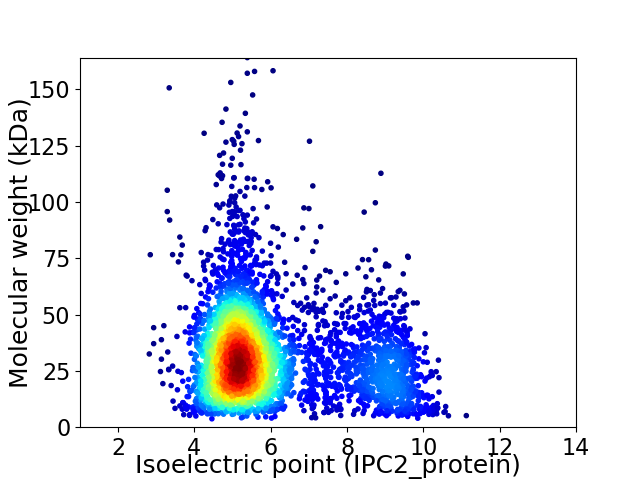
Virtual 2D-PAGE plot for 4197 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I3TEJ0|A0A1I3TEJ0_9RHOB ABC-2 type transport system permease protein OS=Celeribacter neptunius OX=588602 GN=SAMN04487991_2693 PE=4 SV=1
MM1 pKa = 7.19SAIEE5 pKa = 3.99FSEE8 pKa = 4.09NGTAVMFCSHH18 pKa = 7.08ILSGGRR24 pKa = 11.84MTKK27 pKa = 10.56YY28 pKa = 9.71KK29 pKa = 11.16GEE31 pKa = 4.23VASCAPALVRR41 pKa = 11.84PSVVFPRR48 pKa = 11.84LFSAPTVSALALAAGLSVAPVLLPSEE74 pKa = 4.28ITGGAGAAWAACSPSNAGTAGDD96 pKa = 5.84DD97 pKa = 5.2IILCDD102 pKa = 3.99AANDD106 pKa = 4.07PGGANVSGFTGNDD119 pKa = 3.6TITLDD124 pKa = 4.06GMVTVGNISGGDD136 pKa = 3.53GVDD139 pKa = 3.93TINLLSGTVINRR151 pKa = 11.84VVWGNGDD158 pKa = 3.34EE159 pKa = 4.39SQILDD164 pKa = 3.84LTNVTLAGDD173 pKa = 3.68LRR175 pKa = 11.84FGGGSDD181 pKa = 3.44NFTLVAGSVGSNVFLGDD198 pKa = 3.89LEE200 pKa = 4.49NFGTGVDD207 pKa = 4.77DD208 pKa = 5.26VFTLDD213 pKa = 3.63GGHH216 pKa = 5.89VAWEE220 pKa = 3.78ISGDD224 pKa = 3.66QDD226 pKa = 3.58RR227 pKa = 11.84DD228 pKa = 3.75TINIYY233 pKa = 10.24SGSVGRR239 pKa = 11.84RR240 pKa = 11.84IKK242 pKa = 10.82GYY244 pKa = 10.84DD245 pKa = 3.44GDD247 pKa = 6.0DD248 pKa = 3.69VINWSGGTIGIGVYY262 pKa = 10.86GDD264 pKa = 3.68GDD266 pKa = 3.79WYY268 pKa = 10.62DD269 pKa = 3.73GLDD272 pKa = 3.27WFPTYY277 pKa = 10.92GSDD280 pKa = 3.22TVTVSASEE288 pKa = 4.01YY289 pKa = 10.8DD290 pKa = 3.57GTQMLDD296 pKa = 3.46GGDD299 pKa = 3.76DD300 pKa = 3.98AYY302 pKa = 10.11TSDD305 pKa = 4.86GYY307 pKa = 11.71SDD309 pKa = 3.6TLNLNGLTVSANGTYY324 pKa = 9.58IVNWEE329 pKa = 4.2VANLDD334 pKa = 3.46ATALTISDD342 pKa = 4.2GLWDD346 pKa = 3.78VGIDD350 pKa = 3.4GATGSGVFLTNGSEE364 pKa = 4.68LEE366 pKa = 4.18ALDD369 pKa = 5.19ALTLNGNLQIDD380 pKa = 4.26GTSQFTGTGAGAGVYY395 pKa = 9.93NINGTVLNSGTITTYY410 pKa = 11.11DD411 pKa = 3.47GVTGDD416 pKa = 3.9VLTISGDD423 pKa = 3.65YY424 pKa = 11.12SGDD427 pKa = 3.53GQLLFDD433 pKa = 5.55ADD435 pKa = 4.31LASGTGDD442 pKa = 3.45ALVIDD447 pKa = 4.66GDD449 pKa = 4.23VTGGTTTIMVNDD461 pKa = 3.92VTSGPISGNDD471 pKa = 3.24VVLVDD476 pKa = 3.59VTGATAEE483 pKa = 4.29GDD485 pKa = 3.71FVLAGGPLTVGAYY498 pKa = 10.03DD499 pKa = 3.71YY500 pKa = 11.65DD501 pKa = 5.37LIYY504 pKa = 10.91QPGTFVLAGSLNSIGAMYY522 pKa = 9.58EE523 pKa = 3.79AAPLILAGFNRR534 pKa = 11.84LPTLEE539 pKa = 3.9QRR541 pKa = 11.84VGEE544 pKa = 4.21RR545 pKa = 11.84EE546 pKa = 4.09LTGSDD551 pKa = 3.21TASGSWIRR559 pKa = 11.84FYY561 pKa = 11.56GDD563 pKa = 3.14SSDD566 pKa = 3.4MATNSGSNISDD577 pKa = 3.64HH578 pKa = 6.98RR579 pKa = 11.84WGLQAGWDD587 pKa = 3.68EE588 pKa = 3.98RR589 pKa = 11.84LEE591 pKa = 4.18PSEE594 pKa = 3.99TGQWVIGVTAQYY606 pKa = 7.98GTQNASVSMASGAGKK621 pKa = 10.37VSSSNYY627 pKa = 10.09GIGATATWYY636 pKa = 11.06GNDD639 pKa = 3.65GLYY642 pKa = 10.71ADD644 pKa = 4.92LQGQVSWISSDD655 pKa = 3.56FSSSSVGVLATGEE668 pKa = 4.1RR669 pKa = 11.84ANAYY673 pKa = 8.67AASIEE678 pKa = 4.15TGYY681 pKa = 10.96RR682 pKa = 11.84FALDD686 pKa = 3.06QSRR689 pKa = 11.84ALVPQAQMSQGYY701 pKa = 9.55VGGAAFTDD709 pKa = 3.9SQGNAIDD716 pKa = 4.58LGSSSGLTGRR726 pKa = 11.84LGLAYY731 pKa = 9.63EE732 pKa = 4.43YY733 pKa = 10.77EE734 pKa = 4.48RR735 pKa = 11.84SDD737 pKa = 4.0DD738 pKa = 3.74AAGGHH743 pKa = 6.32AKK745 pKa = 10.34LYY747 pKa = 10.83AIGNLLHH754 pKa = 7.12DD755 pKa = 4.75FSEE758 pKa = 5.26GSTVQVAGLDD768 pKa = 3.99LNEE771 pKa = 3.93GTARR775 pKa = 11.84NWGEE779 pKa = 3.84VGIGTSFASDD789 pKa = 4.02EE790 pKa = 3.96NTMFYY795 pKa = 11.36GEE797 pKa = 3.92ASYY800 pKa = 8.64RR801 pKa = 11.84TALDD805 pKa = 4.34GPDD808 pKa = 3.22GTNNGFSATAGLRR821 pKa = 11.84INWW824 pKa = 3.54
MM1 pKa = 7.19SAIEE5 pKa = 3.99FSEE8 pKa = 4.09NGTAVMFCSHH18 pKa = 7.08ILSGGRR24 pKa = 11.84MTKK27 pKa = 10.56YY28 pKa = 9.71KK29 pKa = 11.16GEE31 pKa = 4.23VASCAPALVRR41 pKa = 11.84PSVVFPRR48 pKa = 11.84LFSAPTVSALALAAGLSVAPVLLPSEE74 pKa = 4.28ITGGAGAAWAACSPSNAGTAGDD96 pKa = 5.84DD97 pKa = 5.2IILCDD102 pKa = 3.99AANDD106 pKa = 4.07PGGANVSGFTGNDD119 pKa = 3.6TITLDD124 pKa = 4.06GMVTVGNISGGDD136 pKa = 3.53GVDD139 pKa = 3.93TINLLSGTVINRR151 pKa = 11.84VVWGNGDD158 pKa = 3.34EE159 pKa = 4.39SQILDD164 pKa = 3.84LTNVTLAGDD173 pKa = 3.68LRR175 pKa = 11.84FGGGSDD181 pKa = 3.44NFTLVAGSVGSNVFLGDD198 pKa = 3.89LEE200 pKa = 4.49NFGTGVDD207 pKa = 4.77DD208 pKa = 5.26VFTLDD213 pKa = 3.63GGHH216 pKa = 5.89VAWEE220 pKa = 3.78ISGDD224 pKa = 3.66QDD226 pKa = 3.58RR227 pKa = 11.84DD228 pKa = 3.75TINIYY233 pKa = 10.24SGSVGRR239 pKa = 11.84RR240 pKa = 11.84IKK242 pKa = 10.82GYY244 pKa = 10.84DD245 pKa = 3.44GDD247 pKa = 6.0DD248 pKa = 3.69VINWSGGTIGIGVYY262 pKa = 10.86GDD264 pKa = 3.68GDD266 pKa = 3.79WYY268 pKa = 10.62DD269 pKa = 3.73GLDD272 pKa = 3.27WFPTYY277 pKa = 10.92GSDD280 pKa = 3.22TVTVSASEE288 pKa = 4.01YY289 pKa = 10.8DD290 pKa = 3.57GTQMLDD296 pKa = 3.46GGDD299 pKa = 3.76DD300 pKa = 3.98AYY302 pKa = 10.11TSDD305 pKa = 4.86GYY307 pKa = 11.71SDD309 pKa = 3.6TLNLNGLTVSANGTYY324 pKa = 9.58IVNWEE329 pKa = 4.2VANLDD334 pKa = 3.46ATALTISDD342 pKa = 4.2GLWDD346 pKa = 3.78VGIDD350 pKa = 3.4GATGSGVFLTNGSEE364 pKa = 4.68LEE366 pKa = 4.18ALDD369 pKa = 5.19ALTLNGNLQIDD380 pKa = 4.26GTSQFTGTGAGAGVYY395 pKa = 9.93NINGTVLNSGTITTYY410 pKa = 11.11DD411 pKa = 3.47GVTGDD416 pKa = 3.9VLTISGDD423 pKa = 3.65YY424 pKa = 11.12SGDD427 pKa = 3.53GQLLFDD433 pKa = 5.55ADD435 pKa = 4.31LASGTGDD442 pKa = 3.45ALVIDD447 pKa = 4.66GDD449 pKa = 4.23VTGGTTTIMVNDD461 pKa = 3.92VTSGPISGNDD471 pKa = 3.24VVLVDD476 pKa = 3.59VTGATAEE483 pKa = 4.29GDD485 pKa = 3.71FVLAGGPLTVGAYY498 pKa = 10.03DD499 pKa = 3.71YY500 pKa = 11.65DD501 pKa = 5.37LIYY504 pKa = 10.91QPGTFVLAGSLNSIGAMYY522 pKa = 9.58EE523 pKa = 3.79AAPLILAGFNRR534 pKa = 11.84LPTLEE539 pKa = 3.9QRR541 pKa = 11.84VGEE544 pKa = 4.21RR545 pKa = 11.84EE546 pKa = 4.09LTGSDD551 pKa = 3.21TASGSWIRR559 pKa = 11.84FYY561 pKa = 11.56GDD563 pKa = 3.14SSDD566 pKa = 3.4MATNSGSNISDD577 pKa = 3.64HH578 pKa = 6.98RR579 pKa = 11.84WGLQAGWDD587 pKa = 3.68EE588 pKa = 3.98RR589 pKa = 11.84LEE591 pKa = 4.18PSEE594 pKa = 3.99TGQWVIGVTAQYY606 pKa = 7.98GTQNASVSMASGAGKK621 pKa = 10.37VSSSNYY627 pKa = 10.09GIGATATWYY636 pKa = 11.06GNDD639 pKa = 3.65GLYY642 pKa = 10.71ADD644 pKa = 4.92LQGQVSWISSDD655 pKa = 3.56FSSSSVGVLATGEE668 pKa = 4.1RR669 pKa = 11.84ANAYY673 pKa = 8.67AASIEE678 pKa = 4.15TGYY681 pKa = 10.96RR682 pKa = 11.84FALDD686 pKa = 3.06QSRR689 pKa = 11.84ALVPQAQMSQGYY701 pKa = 9.55VGGAAFTDD709 pKa = 3.9SQGNAIDD716 pKa = 4.58LGSSSGLTGRR726 pKa = 11.84LGLAYY731 pKa = 9.63EE732 pKa = 4.43YY733 pKa = 10.77EE734 pKa = 4.48RR735 pKa = 11.84SDD737 pKa = 4.0DD738 pKa = 3.74AAGGHH743 pKa = 6.32AKK745 pKa = 10.34LYY747 pKa = 10.83AIGNLLHH754 pKa = 7.12DD755 pKa = 4.75FSEE758 pKa = 5.26GSTVQVAGLDD768 pKa = 3.99LNEE771 pKa = 3.93GTARR775 pKa = 11.84NWGEE779 pKa = 3.84VGIGTSFASDD789 pKa = 4.02EE790 pKa = 3.96NTMFYY795 pKa = 11.36GEE797 pKa = 3.92ASYY800 pKa = 8.64RR801 pKa = 11.84TALDD805 pKa = 4.34GPDD808 pKa = 3.22GTNNGFSATAGLRR821 pKa = 11.84INWW824 pKa = 3.54
Molecular weight: 84.44 kDa
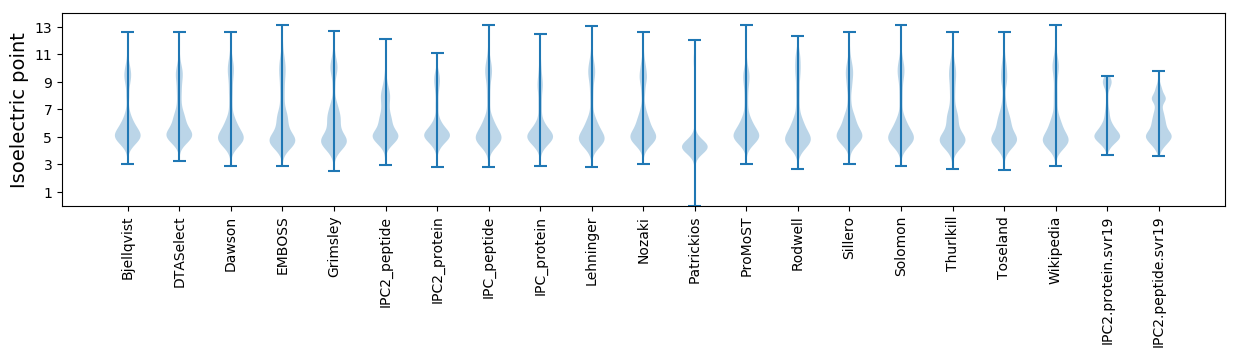
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I3RB13|A0A1I3RB13_9RHOB Nitrous oxidase accessory protein OS=Celeribacter neptunius OX=588602 GN=SAMN04487991_2114 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.44AGRR28 pKa = 11.84KK29 pKa = 8.46VLNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
MM1 pKa = 7.35KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.31QPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.42GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.44AGRR28 pKa = 11.84KK29 pKa = 8.46VLNARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.07SLSAA44 pKa = 3.93
Molecular weight: 5.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1294092 |
32 |
1511 |
308.3 |
33.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.893 ± 0.05 | 0.865 ± 0.012 |
5.882 ± 0.035 | 6.389 ± 0.037 |
3.841 ± 0.025 | 8.584 ± 0.036 |
2.073 ± 0.017 | 5.292 ± 0.026 |
3.553 ± 0.027 | 10.17 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.891 ± 0.02 | 2.557 ± 0.018 |
5.001 ± 0.026 | 3.088 ± 0.021 |
6.279 ± 0.035 | 5.399 ± 0.025 |
5.522 ± 0.03 | 7.066 ± 0.026 |
1.343 ± 0.015 | 2.312 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
