
Hubei picorna-like virus 71
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
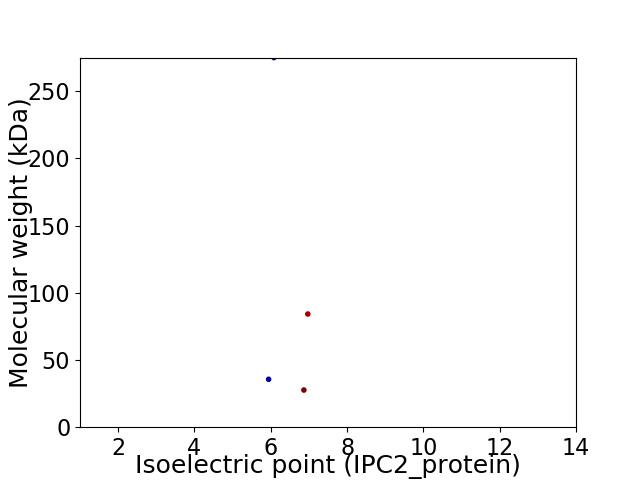
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KJ91|A0A1L3KJ91_9VIRU Uncharacterized protein OS=Hubei picorna-like virus 71 OX=1923155 PE=4 SV=1
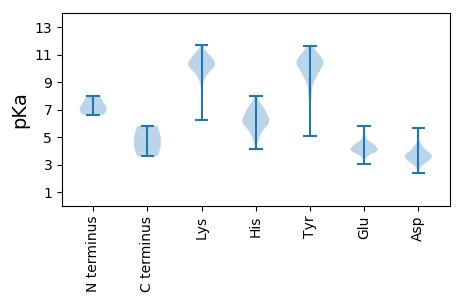
MM1 pKa = 7.95ASACKK6 pKa = 9.87NCSPICQGVRR16 pKa = 11.84NCVTTTIEE24 pKa = 4.07SLRR27 pKa = 11.84TNLTALSFVSPTSMQAITALRR48 pKa = 11.84DD49 pKa = 3.45SLLVAKK55 pKa = 10.4GCIEE59 pKa = 3.86SSDD62 pKa = 3.54YY63 pKa = 11.07GRR65 pKa = 11.84ARR67 pKa = 11.84FVLTAGTPTAKK78 pKa = 9.95EE79 pKa = 3.89LSGLPSNTPFIRR91 pKa = 11.84ALAVAIGRR99 pKa = 11.84SYY101 pKa = 11.51SAIQTNAIDD110 pKa = 4.2TLQKK114 pKa = 9.91ICPNEE119 pKa = 3.85QHH121 pKa = 6.02HH122 pKa = 7.23HH123 pKa = 6.13IMYY126 pKa = 10.24QPNDD130 pKa = 4.11DD131 pKa = 4.89PLSCTPWKK139 pKa = 10.04FRR141 pKa = 11.84RR142 pKa = 11.84LIQAGTVDD150 pKa = 3.75RR151 pKa = 11.84ATADD155 pKa = 3.29VFEE158 pKa = 4.55GFEE161 pKa = 4.59NIFLLHH167 pKa = 7.03EE168 pKa = 4.67DD169 pKa = 3.42PGYY172 pKa = 10.53EE173 pKa = 3.91GHH175 pKa = 7.36HH176 pKa = 6.74NITDD180 pKa = 3.87TLRR183 pKa = 11.84TCHH186 pKa = 5.72GTHH189 pKa = 7.08DD190 pKa = 3.55RR191 pKa = 11.84CLYY194 pKa = 10.28CVLGYY199 pKa = 8.69KK200 pKa = 9.5HH201 pKa = 7.01CYY203 pKa = 9.0ASRR206 pKa = 11.84SHH208 pKa = 6.77FVRR211 pKa = 11.84VCGNDD216 pKa = 3.03EE217 pKa = 3.94CAGCYY222 pKa = 9.65FIEE225 pKa = 4.22HH226 pKa = 6.58HH227 pKa = 6.37VLGRR231 pKa = 11.84DD232 pKa = 3.49VNHH235 pKa = 6.21TRR237 pKa = 11.84MIQFYY242 pKa = 10.27KK243 pKa = 9.57FAQMYY248 pKa = 9.02HH249 pKa = 6.61FYY251 pKa = 11.25ANRR254 pKa = 11.84HH255 pKa = 4.14YY256 pKa = 10.96QIDD259 pKa = 4.26EE260 pKa = 4.2DD261 pKa = 4.32KK262 pKa = 11.08MPIPEE267 pKa = 4.38VNTYY271 pKa = 10.5SYY273 pKa = 11.38PLFTYY278 pKa = 9.75RR279 pKa = 11.84HH280 pKa = 5.31TRR282 pKa = 11.84TLYY285 pKa = 7.92TTEE288 pKa = 3.88SQDD291 pKa = 4.09RR292 pKa = 11.84MIEE295 pKa = 3.8NAMNEE300 pKa = 4.02DD301 pKa = 5.33GEE303 pKa = 4.7IEE305 pKa = 3.94WHH307 pKa = 6.56RR308 pKa = 11.84LAEE311 pKa = 4.48SII313 pKa = 5.03
MM1 pKa = 7.95ASACKK6 pKa = 9.87NCSPICQGVRR16 pKa = 11.84NCVTTTIEE24 pKa = 4.07SLRR27 pKa = 11.84TNLTALSFVSPTSMQAITALRR48 pKa = 11.84DD49 pKa = 3.45SLLVAKK55 pKa = 10.4GCIEE59 pKa = 3.86SSDD62 pKa = 3.54YY63 pKa = 11.07GRR65 pKa = 11.84ARR67 pKa = 11.84FVLTAGTPTAKK78 pKa = 9.95EE79 pKa = 3.89LSGLPSNTPFIRR91 pKa = 11.84ALAVAIGRR99 pKa = 11.84SYY101 pKa = 11.51SAIQTNAIDD110 pKa = 4.2TLQKK114 pKa = 9.91ICPNEE119 pKa = 3.85QHH121 pKa = 6.02HH122 pKa = 7.23HH123 pKa = 6.13IMYY126 pKa = 10.24QPNDD130 pKa = 4.11DD131 pKa = 4.89PLSCTPWKK139 pKa = 10.04FRR141 pKa = 11.84RR142 pKa = 11.84LIQAGTVDD150 pKa = 3.75RR151 pKa = 11.84ATADD155 pKa = 3.29VFEE158 pKa = 4.55GFEE161 pKa = 4.59NIFLLHH167 pKa = 7.03EE168 pKa = 4.67DD169 pKa = 3.42PGYY172 pKa = 10.53EE173 pKa = 3.91GHH175 pKa = 7.36HH176 pKa = 6.74NITDD180 pKa = 3.87TLRR183 pKa = 11.84TCHH186 pKa = 5.72GTHH189 pKa = 7.08DD190 pKa = 3.55RR191 pKa = 11.84CLYY194 pKa = 10.28CVLGYY199 pKa = 8.69KK200 pKa = 9.5HH201 pKa = 7.01CYY203 pKa = 9.0ASRR206 pKa = 11.84SHH208 pKa = 6.77FVRR211 pKa = 11.84VCGNDD216 pKa = 3.03EE217 pKa = 3.94CAGCYY222 pKa = 9.65FIEE225 pKa = 4.22HH226 pKa = 6.58HH227 pKa = 6.37VLGRR231 pKa = 11.84DD232 pKa = 3.49VNHH235 pKa = 6.21TRR237 pKa = 11.84MIQFYY242 pKa = 10.27KK243 pKa = 9.57FAQMYY248 pKa = 9.02HH249 pKa = 6.61FYY251 pKa = 11.25ANRR254 pKa = 11.84HH255 pKa = 4.14YY256 pKa = 10.96QIDD259 pKa = 4.26EE260 pKa = 4.2DD261 pKa = 4.32KK262 pKa = 11.08MPIPEE267 pKa = 4.38VNTYY271 pKa = 10.5SYY273 pKa = 11.38PLFTYY278 pKa = 9.75RR279 pKa = 11.84HH280 pKa = 5.31TRR282 pKa = 11.84TLYY285 pKa = 7.92TTEE288 pKa = 3.88SQDD291 pKa = 4.09RR292 pKa = 11.84MIEE295 pKa = 3.8NAMNEE300 pKa = 4.02DD301 pKa = 5.33GEE303 pKa = 4.7IEE305 pKa = 3.94WHH307 pKa = 6.56RR308 pKa = 11.84LAEE311 pKa = 4.48SII313 pKa = 5.03
Molecular weight: 35.67 kDa
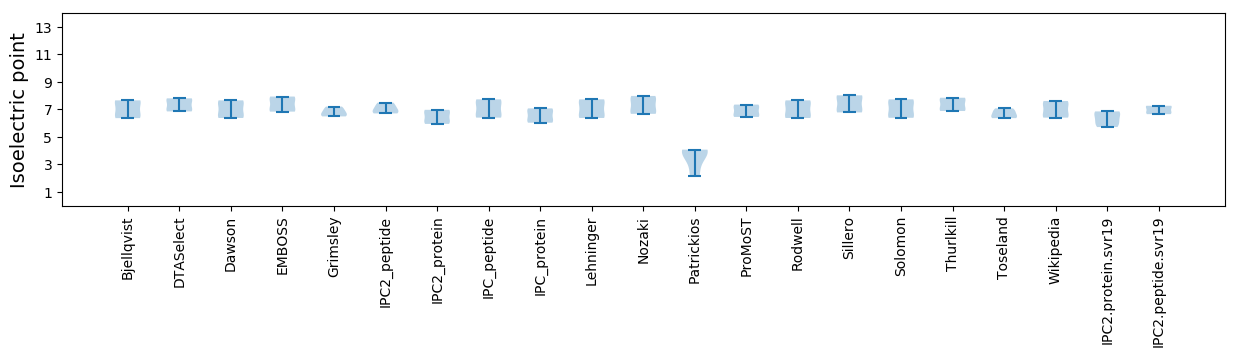
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJ89|A0A1L3KJ89_9VIRU Calici_coat domain-containing protein OS=Hubei picorna-like virus 71 OX=1923155 PE=4 SV=1
MM1 pKa = 6.63NTKK4 pKa = 8.67TLQWHH9 pKa = 5.11PRR11 pKa = 11.84ARR13 pKa = 11.84IVRR16 pKa = 11.84QSVRR20 pKa = 11.84GYY22 pKa = 8.6VTVSPLPLNHH32 pKa = 6.54SEE34 pKa = 4.57RR35 pKa = 11.84TLQLCPLCLQQVCKK49 pKa = 10.46LLQHH53 pKa = 6.62SEE55 pKa = 4.04TLYY58 pKa = 11.07LLQKK62 pKa = 10.38GALNRR67 pKa = 11.84AIMVEE72 pKa = 4.15LGSYY76 pKa = 10.43LPLEE80 pKa = 4.27PQRR83 pKa = 11.84PRR85 pKa = 11.84NCQGYY90 pKa = 8.53LAILPLYY97 pKa = 10.39AHH99 pKa = 7.33LPLQSDD105 pKa = 3.71DD106 pKa = 3.39HH107 pKa = 5.87TVPFRR112 pKa = 11.84RR113 pKa = 11.84MLSTHH118 pKa = 4.57FRR120 pKa = 11.84KK121 pKa = 9.69YY122 pKa = 11.16VPMNSIITLCTNPMMTHH139 pKa = 5.37FHH141 pKa = 6.06AHH143 pKa = 7.08LGNSDD148 pKa = 3.38DD149 pKa = 4.99SSRR152 pKa = 11.84LEE154 pKa = 3.67QWIEE158 pKa = 3.81QQQMCLKK165 pKa = 10.58DD166 pKa = 3.6SRR168 pKa = 11.84TFSYY172 pKa = 11.05YY173 pKa = 10.27MKK175 pKa = 9.86IQDD178 pKa = 4.06MKK180 pKa = 11.04AITISQIPSALATGPTIDD198 pKa = 4.53AYY200 pKa = 9.65TVSSDD205 pKa = 3.82INTAMQAAATLLEE218 pKa = 4.49FAEE221 pKa = 4.47TMNAQAATSSNTMCLEE237 pKa = 4.2EE238 pKa = 4.11TSTTQEE244 pKa = 3.59
MM1 pKa = 6.63NTKK4 pKa = 8.67TLQWHH9 pKa = 5.11PRR11 pKa = 11.84ARR13 pKa = 11.84IVRR16 pKa = 11.84QSVRR20 pKa = 11.84GYY22 pKa = 8.6VTVSPLPLNHH32 pKa = 6.54SEE34 pKa = 4.57RR35 pKa = 11.84TLQLCPLCLQQVCKK49 pKa = 10.46LLQHH53 pKa = 6.62SEE55 pKa = 4.04TLYY58 pKa = 11.07LLQKK62 pKa = 10.38GALNRR67 pKa = 11.84AIMVEE72 pKa = 4.15LGSYY76 pKa = 10.43LPLEE80 pKa = 4.27PQRR83 pKa = 11.84PRR85 pKa = 11.84NCQGYY90 pKa = 8.53LAILPLYY97 pKa = 10.39AHH99 pKa = 7.33LPLQSDD105 pKa = 3.71DD106 pKa = 3.39HH107 pKa = 5.87TVPFRR112 pKa = 11.84RR113 pKa = 11.84MLSTHH118 pKa = 4.57FRR120 pKa = 11.84KK121 pKa = 9.69YY122 pKa = 11.16VPMNSIITLCTNPMMTHH139 pKa = 5.37FHH141 pKa = 6.06AHH143 pKa = 7.08LGNSDD148 pKa = 3.38DD149 pKa = 4.99SSRR152 pKa = 11.84LEE154 pKa = 3.67QWIEE158 pKa = 3.81QQQMCLKK165 pKa = 10.58DD166 pKa = 3.6SRR168 pKa = 11.84TFSYY172 pKa = 11.05YY173 pKa = 10.27MKK175 pKa = 9.86IQDD178 pKa = 4.06MKK180 pKa = 11.04AITISQIPSALATGPTIDD198 pKa = 4.53AYY200 pKa = 9.65TVSSDD205 pKa = 3.82INTAMQAAATLLEE218 pKa = 4.49FAEE221 pKa = 4.47TMNAQAATSSNTMCLEE237 pKa = 4.2EE238 pKa = 4.11TSTTQEE244 pKa = 3.59
Molecular weight: 27.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3725 |
244 |
2408 |
931.3 |
105.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.738 ± 0.6 | 2.013 ± 0.549 |
5.342 ± 0.595 | 6.013 ± 0.801 |
4.268 ± 0.45 | 5.53 ± 0.597 |
2.819 ± 0.543 | 5.691 ± 0.408 |
6.282 ± 1.429 | 7.705 ± 0.756 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.872 ± 0.315 | 5.477 ± 0.746 |
5.074 ± 0.52 | 4.591 ± 0.805 |
4.537 ± 0.454 | 6.577 ± 0.568 |
6.899 ± 0.76 | 5.718 ± 0.744 |
1.342 ± 0.216 | 4.51 ± 0.155 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
