
Fusarium flagelliforme
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina;
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
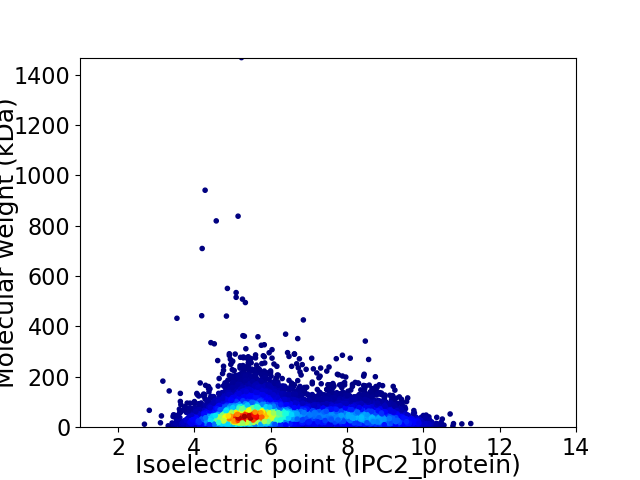
Virtual 2D-PAGE plot for 13039 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
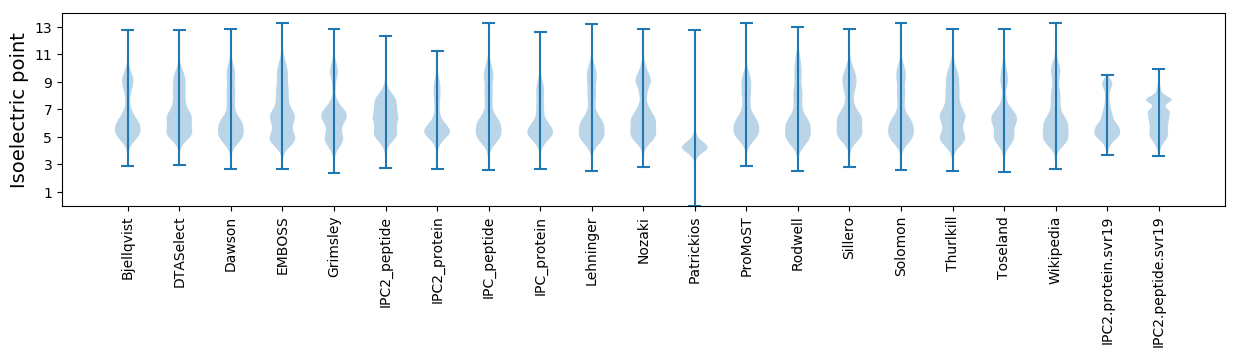
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A395MWI9|A0A395MWI9_9HYPO Uncharacterized protein OS=Fusarium flagelliforme OX=2675880 GN=FIE12Z_3652 PE=4 SV=1
MM1 pKa = 7.91RR2 pKa = 11.84ISASLGVLVATGLLGHH18 pKa = 6.34SASQATDD25 pKa = 3.23YY26 pKa = 11.54GNNNGSPVGNQPGASGNGIPGNNGGASSSMPYY58 pKa = 9.81PSNAVGSVPGSMPSGVGPVGPVSPGSNGLPPTVPGGIPGNLPGSTSPGDD107 pKa = 3.59ASNGYY112 pKa = 8.52PPNGPNGSPGNFPGPVSPGGGSNSLPPNGPGGVPGSIPGSIPGNSPINGPGNVPGNVPGNAPGNVPGGVPGAGSVIVPGSFPTNIPQNVPGAPGSPDD209 pKa = 3.11SDD211 pKa = 3.19ASNVPTCPLRR221 pKa = 11.84STKK224 pKa = 9.97TVVVTVYY231 pKa = 9.76STGSQNEE238 pKa = 4.27SDD240 pKa = 5.28FPWPDD245 pKa = 3.59DD246 pKa = 3.87SDD248 pKa = 4.67SDD250 pKa = 4.13GSSTPTPMPTSPPGATPWIPTSGIPIFTTLTLDD283 pKa = 2.48IWGPDD288 pKa = 3.49GSPSNSGSDD297 pKa = 3.83SNDD300 pKa = 3.05GTSGNGNNPSGSGSPGDD317 pKa = 3.67GSGGNDD323 pKa = 3.51GANSGSGNGSDD334 pKa = 4.81SDD336 pKa = 5.15GDD338 pKa = 3.88DD339 pKa = 3.5SQNNPGSPFPFPASGPNGAPQYY361 pKa = 10.25SDD363 pKa = 3.02NRR365 pKa = 11.84FGGSSAGIPGATGDD379 pKa = 3.56NTYY382 pKa = 10.68VHH384 pKa = 6.9TPLQTPVSPNGGVTGNQPGLPSTGPAQEE412 pKa = 4.95ASDD415 pKa = 4.18SLPVTTQAGPEE426 pKa = 4.25GQGEE430 pKa = 4.17NSPWLTNVPGGNGASQPSPSYY451 pKa = 9.91ITITGQDD458 pKa = 3.7GLPTVVSNGGNMIPGQAQPSANSGFPPNGNNPGLPVGGSSGFPLPPPAITGTPGSQNPSNSPAGSLPGSGPDD530 pKa = 4.2EE531 pKa = 5.46GITTCATFTITGTDD545 pKa = 3.65GLPTVVDD552 pKa = 4.1TTWVIPGTFDD562 pKa = 3.25PQAPLSTITSEE573 pKa = 4.36LGLPTFPFTIPGSPGDD589 pKa = 4.0ASGHH593 pKa = 5.17SAMTTSTSYY602 pKa = 11.2TMIGADD608 pKa = 3.6GNPTVVHH615 pKa = 5.2TTWTIPTAEE624 pKa = 4.81AGFGDD629 pKa = 4.63PGDD632 pKa = 3.82KK633 pKa = 10.42SALVSSGDD641 pKa = 3.4FMTVTEE647 pKa = 5.04LPPFGSSGSGGDD659 pKa = 3.74NPTAPNSDD667 pKa = 3.39NASPITTCTSYY678 pKa = 10.59TVIGTDD684 pKa = 3.59GLPTVVDD691 pKa = 3.73STFVIPGPVNTGATAGGDD709 pKa = 3.52PQVPSGASDD718 pKa = 3.82ALPNGATSPATIQITSSPGLTPPAPTDD745 pKa = 3.35GTGIGSIGNGDD756 pKa = 3.06ITTCTSYY763 pKa = 10.86TITGPDD769 pKa = 3.89GLPTIVDD776 pKa = 4.18STWVIPGPANTQSEE790 pKa = 4.82LPGNPSFVSDD800 pKa = 5.03SIPSGLPTGFPGPMTASAGIPSDD823 pKa = 4.53DD824 pKa = 4.07NQGNMPGVTTCITYY838 pKa = 7.52TTIGSDD844 pKa = 3.66GLPTVVDD851 pKa = 3.73TTFVIPVATATPAGTSLNVNGNGQTGSSPNPTGAFTTFTTAIVLGPDD898 pKa = 3.76GNPTSTVQTIVFSDD912 pKa = 3.83SSALGVSAGPPQGATSGVTPLGPSGPVFTTGDD944 pKa = 4.03SNSLPTDD951 pKa = 3.6TPSLNGYY958 pKa = 10.3DD959 pKa = 4.19DD960 pKa = 5.97GIPGASISVVLTGGSAAAQGTSVSGTTTGTLTWTVTSITNPAGGPVFSQGAGQPPFASGPSDD1022 pKa = 3.27NSASDD1027 pKa = 4.01GMAQPAYY1034 pKa = 10.14GSVATEE1040 pKa = 3.8STLFPLSAIQTSTWTNVIKK1059 pKa = 10.92AEE1061 pKa = 4.24TTSYY1065 pKa = 9.33TFNYY1069 pKa = 9.4PVTTLATVNVPKK1081 pKa = 10.56KK1082 pKa = 10.13RR1083 pKa = 11.84FARR1086 pKa = 11.84RR1087 pKa = 11.84QEE1089 pKa = 3.54MTAWSNSTTSASSAISASTSTTSEE1113 pKa = 3.72ASSPSICPAGGNIGNTTIDD1132 pKa = 3.86FDD1134 pKa = 4.06TSKK1137 pKa = 10.53PGPLFNPVEE1146 pKa = 4.53NIWFSGGFLIAPPATQQSQPYY1167 pKa = 8.49IPSSGGQLVEE1177 pKa = 4.92FVPPALSNTTTTISGDD1193 pKa = 3.29VAQIGVGPHH1202 pKa = 6.14AASPCFRR1209 pKa = 11.84FDD1211 pKa = 3.51FFGAEE1216 pKa = 4.46LGCDD1220 pKa = 3.36AQGDD1224 pKa = 4.26EE1225 pKa = 4.8EE1226 pKa = 4.43WCEE1229 pKa = 4.02FDD1231 pKa = 3.62ISAYY1235 pKa = 9.59RR1236 pKa = 11.84WNEE1239 pKa = 3.59TSSTEE1244 pKa = 4.22EE1245 pKa = 3.76SIAWSEE1251 pKa = 4.39TKK1253 pKa = 9.86QVPACPKK1260 pKa = 9.5FSEE1263 pKa = 4.6GGYY1266 pKa = 9.82QLTRR1270 pKa = 11.84VDD1272 pKa = 3.71LDD1274 pKa = 3.79GYY1276 pKa = 11.0KK1277 pKa = 10.39DD1278 pKa = 3.66LSSILITLRR1287 pKa = 11.84VSSNLRR1293 pKa = 11.84VWWGDD1298 pKa = 3.26DD1299 pKa = 3.78FRR1301 pKa = 11.84VGWTDD1306 pKa = 3.2NSCVAASCRR1315 pKa = 11.84ANAPSQFVKK1324 pKa = 10.72RR1325 pKa = 11.84EE1326 pKa = 4.09TIISALRR1333 pKa = 11.84QGVYY1337 pKa = 7.64QWTPYY1342 pKa = 9.18EE1343 pKa = 4.43LKK1345 pKa = 10.68RR1346 pKa = 11.84LDD1348 pKa = 5.16DD1349 pKa = 3.85SLVWEE1354 pKa = 4.61SANN1357 pKa = 3.76
MM1 pKa = 7.91RR2 pKa = 11.84ISASLGVLVATGLLGHH18 pKa = 6.34SASQATDD25 pKa = 3.23YY26 pKa = 11.54GNNNGSPVGNQPGASGNGIPGNNGGASSSMPYY58 pKa = 9.81PSNAVGSVPGSMPSGVGPVGPVSPGSNGLPPTVPGGIPGNLPGSTSPGDD107 pKa = 3.59ASNGYY112 pKa = 8.52PPNGPNGSPGNFPGPVSPGGGSNSLPPNGPGGVPGSIPGSIPGNSPINGPGNVPGNVPGNAPGNVPGGVPGAGSVIVPGSFPTNIPQNVPGAPGSPDD209 pKa = 3.11SDD211 pKa = 3.19ASNVPTCPLRR221 pKa = 11.84STKK224 pKa = 9.97TVVVTVYY231 pKa = 9.76STGSQNEE238 pKa = 4.27SDD240 pKa = 5.28FPWPDD245 pKa = 3.59DD246 pKa = 3.87SDD248 pKa = 4.67SDD250 pKa = 4.13GSSTPTPMPTSPPGATPWIPTSGIPIFTTLTLDD283 pKa = 2.48IWGPDD288 pKa = 3.49GSPSNSGSDD297 pKa = 3.83SNDD300 pKa = 3.05GTSGNGNNPSGSGSPGDD317 pKa = 3.67GSGGNDD323 pKa = 3.51GANSGSGNGSDD334 pKa = 4.81SDD336 pKa = 5.15GDD338 pKa = 3.88DD339 pKa = 3.5SQNNPGSPFPFPASGPNGAPQYY361 pKa = 10.25SDD363 pKa = 3.02NRR365 pKa = 11.84FGGSSAGIPGATGDD379 pKa = 3.56NTYY382 pKa = 10.68VHH384 pKa = 6.9TPLQTPVSPNGGVTGNQPGLPSTGPAQEE412 pKa = 4.95ASDD415 pKa = 4.18SLPVTTQAGPEE426 pKa = 4.25GQGEE430 pKa = 4.17NSPWLTNVPGGNGASQPSPSYY451 pKa = 9.91ITITGQDD458 pKa = 3.7GLPTVVSNGGNMIPGQAQPSANSGFPPNGNNPGLPVGGSSGFPLPPPAITGTPGSQNPSNSPAGSLPGSGPDD530 pKa = 4.2EE531 pKa = 5.46GITTCATFTITGTDD545 pKa = 3.65GLPTVVDD552 pKa = 4.1TTWVIPGTFDD562 pKa = 3.25PQAPLSTITSEE573 pKa = 4.36LGLPTFPFTIPGSPGDD589 pKa = 4.0ASGHH593 pKa = 5.17SAMTTSTSYY602 pKa = 11.2TMIGADD608 pKa = 3.6GNPTVVHH615 pKa = 5.2TTWTIPTAEE624 pKa = 4.81AGFGDD629 pKa = 4.63PGDD632 pKa = 3.82KK633 pKa = 10.42SALVSSGDD641 pKa = 3.4FMTVTEE647 pKa = 5.04LPPFGSSGSGGDD659 pKa = 3.74NPTAPNSDD667 pKa = 3.39NASPITTCTSYY678 pKa = 10.59TVIGTDD684 pKa = 3.59GLPTVVDD691 pKa = 3.73STFVIPGPVNTGATAGGDD709 pKa = 3.52PQVPSGASDD718 pKa = 3.82ALPNGATSPATIQITSSPGLTPPAPTDD745 pKa = 3.35GTGIGSIGNGDD756 pKa = 3.06ITTCTSYY763 pKa = 10.86TITGPDD769 pKa = 3.89GLPTIVDD776 pKa = 4.18STWVIPGPANTQSEE790 pKa = 4.82LPGNPSFVSDD800 pKa = 5.03SIPSGLPTGFPGPMTASAGIPSDD823 pKa = 4.53DD824 pKa = 4.07NQGNMPGVTTCITYY838 pKa = 7.52TTIGSDD844 pKa = 3.66GLPTVVDD851 pKa = 3.73TTFVIPVATATPAGTSLNVNGNGQTGSSPNPTGAFTTFTTAIVLGPDD898 pKa = 3.76GNPTSTVQTIVFSDD912 pKa = 3.83SSALGVSAGPPQGATSGVTPLGPSGPVFTTGDD944 pKa = 4.03SNSLPTDD951 pKa = 3.6TPSLNGYY958 pKa = 10.3DD959 pKa = 4.19DD960 pKa = 5.97GIPGASISVVLTGGSAAAQGTSVSGTTTGTLTWTVTSITNPAGGPVFSQGAGQPPFASGPSDD1022 pKa = 3.27NSASDD1027 pKa = 4.01GMAQPAYY1034 pKa = 10.14GSVATEE1040 pKa = 3.8STLFPLSAIQTSTWTNVIKK1059 pKa = 10.92AEE1061 pKa = 4.24TTSYY1065 pKa = 9.33TFNYY1069 pKa = 9.4PVTTLATVNVPKK1081 pKa = 10.56KK1082 pKa = 10.13RR1083 pKa = 11.84FARR1086 pKa = 11.84RR1087 pKa = 11.84QEE1089 pKa = 3.54MTAWSNSTTSASSAISASTSTTSEE1113 pKa = 3.72ASSPSICPAGGNIGNTTIDD1132 pKa = 3.86FDD1134 pKa = 4.06TSKK1137 pKa = 10.53PGPLFNPVEE1146 pKa = 4.53NIWFSGGFLIAPPATQQSQPYY1167 pKa = 8.49IPSSGGQLVEE1177 pKa = 4.92FVPPALSNTTTTISGDD1193 pKa = 3.29VAQIGVGPHH1202 pKa = 6.14AASPCFRR1209 pKa = 11.84FDD1211 pKa = 3.51FFGAEE1216 pKa = 4.46LGCDD1220 pKa = 3.36AQGDD1224 pKa = 4.26EE1225 pKa = 4.8EE1226 pKa = 4.43WCEE1229 pKa = 4.02FDD1231 pKa = 3.62ISAYY1235 pKa = 9.59RR1236 pKa = 11.84WNEE1239 pKa = 3.59TSSTEE1244 pKa = 4.22EE1245 pKa = 3.76SIAWSEE1251 pKa = 4.39TKK1253 pKa = 9.86QVPACPKK1260 pKa = 9.5FSEE1263 pKa = 4.6GGYY1266 pKa = 9.82QLTRR1270 pKa = 11.84VDD1272 pKa = 3.71LDD1274 pKa = 3.79GYY1276 pKa = 11.0KK1277 pKa = 10.39DD1278 pKa = 3.66LSSILITLRR1287 pKa = 11.84VSSNLRR1293 pKa = 11.84VWWGDD1298 pKa = 3.26DD1299 pKa = 3.78FRR1301 pKa = 11.84VGWTDD1306 pKa = 3.2NSCVAASCRR1315 pKa = 11.84ANAPSQFVKK1324 pKa = 10.72RR1325 pKa = 11.84EE1326 pKa = 4.09TIISALRR1333 pKa = 11.84QGVYY1337 pKa = 7.64QWTPYY1342 pKa = 9.18EE1343 pKa = 4.43LKK1345 pKa = 10.68RR1346 pKa = 11.84LDD1348 pKa = 5.16DD1349 pKa = 3.85SLVWEE1354 pKa = 4.61SANN1357 pKa = 3.76
Molecular weight: 134.36 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A395N215|A0A395N215_9HYPO Vacuolar calcium ion transporter OS=Fusarium flagelliforme OX=2675880 GN=FIE12Z_1651 PE=3 SV=1
VV1 pKa = 6.97KK2 pKa = 9.91PRR4 pKa = 11.84PAQSGPRR11 pKa = 11.84PGPPVKK17 pKa = 10.33GGPPRR22 pKa = 11.84GPGPMSPNGGRR33 pKa = 11.84PQSPMGPPRR42 pKa = 11.84GPPGHH47 pKa = 6.63PQGSPRR53 pKa = 11.84GQSPGPFQGPPQGRR67 pKa = 11.84SMSPGPRR74 pKa = 11.84SQSPGPRR81 pKa = 11.84NGPPGGRR88 pKa = 11.84PMSPGPRR95 pKa = 11.84SQSPGPAGGRR105 pKa = 11.84SQSPNGVNRR114 pKa = 11.84RR115 pKa = 11.84NNPPGPSPMNPSQGPTSSGSVNRR138 pKa = 11.84KK139 pKa = 7.96PVPGQAYY146 pKa = 9.86
VV1 pKa = 6.97KK2 pKa = 9.91PRR4 pKa = 11.84PAQSGPRR11 pKa = 11.84PGPPVKK17 pKa = 10.33GGPPRR22 pKa = 11.84GPGPMSPNGGRR33 pKa = 11.84PQSPMGPPRR42 pKa = 11.84GPPGHH47 pKa = 6.63PQGSPRR53 pKa = 11.84GQSPGPFQGPPQGRR67 pKa = 11.84SMSPGPRR74 pKa = 11.84SQSPGPRR81 pKa = 11.84NGPPGGRR88 pKa = 11.84PMSPGPRR95 pKa = 11.84SQSPGPAGGRR105 pKa = 11.84SQSPNGVNRR114 pKa = 11.84RR115 pKa = 11.84NNPPGPSPMNPSQGPTSSGSVNRR138 pKa = 11.84KK139 pKa = 7.96PVPGQAYY146 pKa = 9.86
Molecular weight: 14.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6531987 |
28 |
13355 |
501.0 |
55.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.32 ± 0.019 | 1.268 ± 0.01 |
5.945 ± 0.016 | 6.353 ± 0.024 |
3.757 ± 0.013 | 6.746 ± 0.022 |
2.355 ± 0.011 | 5.061 ± 0.015 |
5.124 ± 0.019 | 8.662 ± 0.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.245 ± 0.008 | 3.837 ± 0.012 |
5.933 ± 0.025 | 4.023 ± 0.014 |
5.783 ± 0.019 | 8.034 ± 0.022 |
6.101 ± 0.028 | 6.094 ± 0.018 |
1.538 ± 0.009 | 2.818 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
