
Hubei toti-like virus 24
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
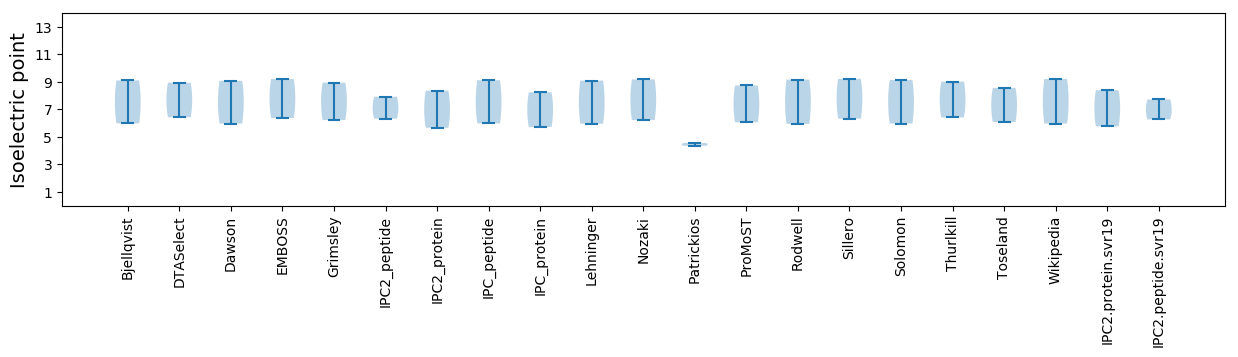
Average proteome isoelectric point is 7.08
Get precalculated fractions of proteins
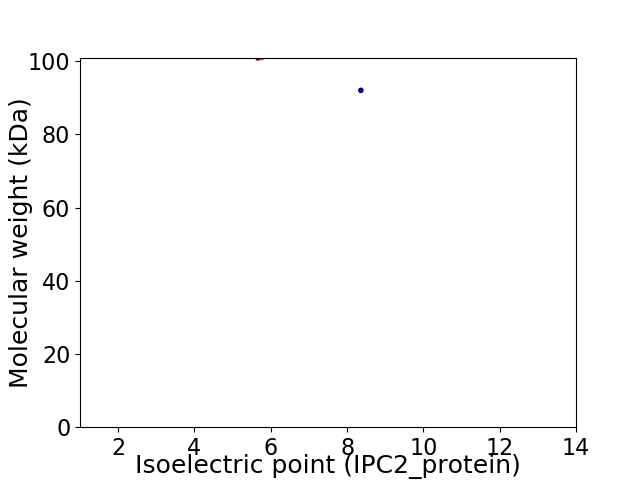
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KFA5|A0A1L3KFA5_9VIRU RdRp catalytic domain-containing protein OS=Hubei toti-like virus 24 OX=1923313 PE=4 SV=1
MM1 pKa = 7.43TIEE4 pKa = 5.34DD5 pKa = 3.91KK6 pKa = 11.03KK7 pKa = 10.78QNCTAITQHH16 pKa = 6.83PLFEE20 pKa = 4.64AAQAAWAQTVPAVLPLNSGNPTHH43 pKa = 7.27CDD45 pKa = 3.09QVAHH49 pKa = 5.19QAIGGAHH56 pKa = 7.17RR57 pKa = 11.84LHH59 pKa = 6.9TYY61 pKa = 7.14WQWAEE66 pKa = 3.84PLRR69 pKa = 11.84LVGVRR74 pKa = 11.84PTTLALGSTILAPHH88 pKa = 6.57LPVRR92 pKa = 11.84VARR95 pKa = 11.84HH96 pKa = 4.81AQRR99 pKa = 11.84IAGPTTDD106 pKa = 5.65LEE108 pKa = 4.5VTPLFQQFAALNNTPVGEE126 pKa = 4.35FQASRR131 pKa = 11.84LFTMXMTKK139 pKa = 9.34DD140 pKa = 3.48TRR142 pKa = 11.84SMALLASVLAQGVSATKK159 pKa = 10.7DD160 pKa = 3.07EE161 pKa = 4.69TPFLRR166 pKa = 11.84AVLISEE172 pKa = 4.43CLIKK176 pKa = 10.53VARR179 pKa = 11.84LPVLTQLPVRR189 pKa = 11.84VAGDD193 pKa = 3.52LRR195 pKa = 11.84LTSVKK200 pKa = 10.27RR201 pKa = 11.84QMVKK205 pKa = 10.6YY206 pKa = 8.95STSAPDD212 pKa = 3.8GNIMAVDD219 pKa = 4.16LARR222 pKa = 11.84FINMYY227 pKa = 10.66ANPSDD232 pKa = 4.17VEE234 pKa = 3.87QHH236 pKa = 5.66FAGFTKK242 pKa = 9.77DD243 pKa = 2.76TWGTTTAVVPFRR255 pKa = 11.84TDD257 pKa = 3.07LSGLRR262 pKa = 11.84SSGPYY267 pKa = 9.56VLSFVDD273 pKa = 3.2TMYY276 pKa = 10.13WNARR280 pKa = 11.84IKK282 pKa = 10.75YY283 pKa = 9.75LLEE286 pKa = 4.49CCPHH290 pKa = 7.2SDD292 pKa = 4.3DD293 pKa = 5.1AEE295 pKa = 4.37KK296 pKa = 11.01VDD298 pKa = 6.89LIAIPNAALGVVDD311 pKa = 5.1GEE313 pKa = 4.41ANILLVLIDD322 pKa = 3.52QVVRR326 pKa = 11.84TNNRR330 pKa = 11.84FEE332 pKa = 4.89LEE334 pKa = 4.04VGGQKK339 pKa = 9.83IPVSGASRR347 pKa = 11.84VTIAPAACSVRR358 pKa = 11.84KK359 pKa = 9.95VVDD362 pKa = 3.53AVFGAGGSAGDD373 pKa = 4.19SPIGQADD380 pKa = 3.96AYY382 pKa = 10.89SAIKK386 pKa = 9.98EE387 pKa = 4.16LIGVIGTSDD396 pKa = 3.77CYY398 pKa = 10.53EE399 pKa = 3.96AALILASEE407 pKa = 4.66MCRR410 pKa = 11.84SFPVDD415 pKa = 3.26PLAKK419 pKa = 9.68VGRR422 pKa = 11.84RR423 pKa = 11.84GDD425 pKa = 4.3SIGLYY430 pKa = 8.37QWEE433 pKa = 4.9EE434 pKa = 3.8DD435 pKa = 3.46TMTNVLGASGPKK447 pKa = 8.19VATWKK452 pKa = 10.51GHH454 pKa = 6.61FDD456 pKa = 4.17DD457 pKa = 5.65GLNKK461 pKa = 10.62SKK463 pKa = 8.11MTYY466 pKa = 8.34LQWYY470 pKa = 9.01AVAQSGMLPEE480 pKa = 5.36AYY482 pKa = 10.16FEE484 pKa = 4.28VGSILTTDD492 pKa = 3.88ASWPVIGSPKK502 pKa = 10.11GGSSLIVNTADD513 pKa = 2.62ITVRR517 pKa = 11.84VARR520 pKa = 11.84ALEE523 pKa = 4.06LVYY526 pKa = 10.65GITTRR531 pKa = 11.84VKK533 pKa = 10.04PDD535 pKa = 3.29TEE537 pKa = 4.44FTLSPWLSASAVLIASQTFAALASAGLSWASLNMLFSVHH576 pKa = 5.55VRR578 pKa = 11.84VPSAVEE584 pKa = 3.91EE585 pKa = 4.34VVSAITNGRR594 pKa = 11.84VTTSPASKK602 pKa = 10.03KK603 pKa = 10.32VIYY606 pKa = 8.88RR607 pKa = 11.84TDD609 pKa = 2.9IAVFSTHH616 pKa = 5.74ATLAATGGQWAADD629 pKa = 3.8DD630 pKa = 4.24SGGFPLPFFLVRR642 pKa = 11.84EE643 pKa = 4.41LVRR646 pKa = 11.84KK647 pKa = 9.62FNVNLVDD654 pKa = 4.56PGHH657 pKa = 7.12LLQEE661 pKa = 4.29RR662 pKa = 11.84MKK664 pKa = 10.87EE665 pKa = 3.88ADD667 pKa = 3.61EE668 pKa = 4.63DD669 pKa = 4.17ADD671 pKa = 4.22IISVCAPVNKK681 pKa = 9.93GWLFVPWYY689 pKa = 8.59TSTVDD694 pKa = 2.97WVRR697 pKa = 11.84RR698 pKa = 11.84PFTVLSTAPKK708 pKa = 8.15GQQIRR713 pKa = 11.84EE714 pKa = 4.08MSVPLYY720 pKa = 8.37WRR722 pKa = 11.84EE723 pKa = 3.68WADD726 pKa = 3.41EE727 pKa = 4.17SSAGMLRR734 pKa = 11.84TRR736 pKa = 11.84NVDD739 pKa = 2.61IPARR743 pKa = 11.84VSLYY747 pKa = 10.25SGMMVDD753 pKa = 5.81QFFADD758 pKa = 3.22QFIRR762 pKa = 11.84QEE764 pKa = 3.83TWSEE768 pKa = 4.23VTSATPVLCHH778 pKa = 6.63HH779 pKa = 7.18GDD781 pKa = 3.38VVADD785 pKa = 3.56VTVPDD790 pKa = 4.63PEE792 pKa = 4.74SPLLRR797 pKa = 11.84AVWSDD802 pKa = 2.72SHH804 pKa = 7.17GFVPPILALEE814 pKa = 4.44APVDD818 pKa = 3.89VDD820 pKa = 4.51GVLPPSGPARR830 pKa = 11.84TNGQRR835 pKa = 11.84LSRR838 pKa = 11.84VLQLPFGASDD848 pKa = 4.01LGRR851 pKa = 11.84ARR853 pKa = 11.84CSPPKK858 pKa = 10.18AEE860 pKa = 4.33SSGTPGLGVNVVPTPRR876 pKa = 11.84RR877 pKa = 11.84EE878 pKa = 4.04LRR880 pKa = 11.84NRR882 pKa = 11.84DD883 pKa = 3.52PLGLLGLNRR892 pKa = 11.84VGGQQAQQPPLNPPPPPAGADD913 pKa = 3.22PALANRR919 pKa = 11.84GAGQPAGAGNDD930 pKa = 3.86DD931 pKa = 3.44QHH933 pKa = 8.56DD934 pKa = 4.02PDD936 pKa = 4.13SRR938 pKa = 3.95
MM1 pKa = 7.43TIEE4 pKa = 5.34DD5 pKa = 3.91KK6 pKa = 11.03KK7 pKa = 10.78QNCTAITQHH16 pKa = 6.83PLFEE20 pKa = 4.64AAQAAWAQTVPAVLPLNSGNPTHH43 pKa = 7.27CDD45 pKa = 3.09QVAHH49 pKa = 5.19QAIGGAHH56 pKa = 7.17RR57 pKa = 11.84LHH59 pKa = 6.9TYY61 pKa = 7.14WQWAEE66 pKa = 3.84PLRR69 pKa = 11.84LVGVRR74 pKa = 11.84PTTLALGSTILAPHH88 pKa = 6.57LPVRR92 pKa = 11.84VARR95 pKa = 11.84HH96 pKa = 4.81AQRR99 pKa = 11.84IAGPTTDD106 pKa = 5.65LEE108 pKa = 4.5VTPLFQQFAALNNTPVGEE126 pKa = 4.35FQASRR131 pKa = 11.84LFTMXMTKK139 pKa = 9.34DD140 pKa = 3.48TRR142 pKa = 11.84SMALLASVLAQGVSATKK159 pKa = 10.7DD160 pKa = 3.07EE161 pKa = 4.69TPFLRR166 pKa = 11.84AVLISEE172 pKa = 4.43CLIKK176 pKa = 10.53VARR179 pKa = 11.84LPVLTQLPVRR189 pKa = 11.84VAGDD193 pKa = 3.52LRR195 pKa = 11.84LTSVKK200 pKa = 10.27RR201 pKa = 11.84QMVKK205 pKa = 10.6YY206 pKa = 8.95STSAPDD212 pKa = 3.8GNIMAVDD219 pKa = 4.16LARR222 pKa = 11.84FINMYY227 pKa = 10.66ANPSDD232 pKa = 4.17VEE234 pKa = 3.87QHH236 pKa = 5.66FAGFTKK242 pKa = 9.77DD243 pKa = 2.76TWGTTTAVVPFRR255 pKa = 11.84TDD257 pKa = 3.07LSGLRR262 pKa = 11.84SSGPYY267 pKa = 9.56VLSFVDD273 pKa = 3.2TMYY276 pKa = 10.13WNARR280 pKa = 11.84IKK282 pKa = 10.75YY283 pKa = 9.75LLEE286 pKa = 4.49CCPHH290 pKa = 7.2SDD292 pKa = 4.3DD293 pKa = 5.1AEE295 pKa = 4.37KK296 pKa = 11.01VDD298 pKa = 6.89LIAIPNAALGVVDD311 pKa = 5.1GEE313 pKa = 4.41ANILLVLIDD322 pKa = 3.52QVVRR326 pKa = 11.84TNNRR330 pKa = 11.84FEE332 pKa = 4.89LEE334 pKa = 4.04VGGQKK339 pKa = 9.83IPVSGASRR347 pKa = 11.84VTIAPAACSVRR358 pKa = 11.84KK359 pKa = 9.95VVDD362 pKa = 3.53AVFGAGGSAGDD373 pKa = 4.19SPIGQADD380 pKa = 3.96AYY382 pKa = 10.89SAIKK386 pKa = 9.98EE387 pKa = 4.16LIGVIGTSDD396 pKa = 3.77CYY398 pKa = 10.53EE399 pKa = 3.96AALILASEE407 pKa = 4.66MCRR410 pKa = 11.84SFPVDD415 pKa = 3.26PLAKK419 pKa = 9.68VGRR422 pKa = 11.84RR423 pKa = 11.84GDD425 pKa = 4.3SIGLYY430 pKa = 8.37QWEE433 pKa = 4.9EE434 pKa = 3.8DD435 pKa = 3.46TMTNVLGASGPKK447 pKa = 8.19VATWKK452 pKa = 10.51GHH454 pKa = 6.61FDD456 pKa = 4.17DD457 pKa = 5.65GLNKK461 pKa = 10.62SKK463 pKa = 8.11MTYY466 pKa = 8.34LQWYY470 pKa = 9.01AVAQSGMLPEE480 pKa = 5.36AYY482 pKa = 10.16FEE484 pKa = 4.28VGSILTTDD492 pKa = 3.88ASWPVIGSPKK502 pKa = 10.11GGSSLIVNTADD513 pKa = 2.62ITVRR517 pKa = 11.84VARR520 pKa = 11.84ALEE523 pKa = 4.06LVYY526 pKa = 10.65GITTRR531 pKa = 11.84VKK533 pKa = 10.04PDD535 pKa = 3.29TEE537 pKa = 4.44FTLSPWLSASAVLIASQTFAALASAGLSWASLNMLFSVHH576 pKa = 5.55VRR578 pKa = 11.84VPSAVEE584 pKa = 3.91EE585 pKa = 4.34VVSAITNGRR594 pKa = 11.84VTTSPASKK602 pKa = 10.03KK603 pKa = 10.32VIYY606 pKa = 8.88RR607 pKa = 11.84TDD609 pKa = 2.9IAVFSTHH616 pKa = 5.74ATLAATGGQWAADD629 pKa = 3.8DD630 pKa = 4.24SGGFPLPFFLVRR642 pKa = 11.84EE643 pKa = 4.41LVRR646 pKa = 11.84KK647 pKa = 9.62FNVNLVDD654 pKa = 4.56PGHH657 pKa = 7.12LLQEE661 pKa = 4.29RR662 pKa = 11.84MKK664 pKa = 10.87EE665 pKa = 3.88ADD667 pKa = 3.61EE668 pKa = 4.63DD669 pKa = 4.17ADD671 pKa = 4.22IISVCAPVNKK681 pKa = 9.93GWLFVPWYY689 pKa = 8.59TSTVDD694 pKa = 2.97WVRR697 pKa = 11.84RR698 pKa = 11.84PFTVLSTAPKK708 pKa = 8.15GQQIRR713 pKa = 11.84EE714 pKa = 4.08MSVPLYY720 pKa = 8.37WRR722 pKa = 11.84EE723 pKa = 3.68WADD726 pKa = 3.41EE727 pKa = 4.17SSAGMLRR734 pKa = 11.84TRR736 pKa = 11.84NVDD739 pKa = 2.61IPARR743 pKa = 11.84VSLYY747 pKa = 10.25SGMMVDD753 pKa = 5.81QFFADD758 pKa = 3.22QFIRR762 pKa = 11.84QEE764 pKa = 3.83TWSEE768 pKa = 4.23VTSATPVLCHH778 pKa = 6.63HH779 pKa = 7.18GDD781 pKa = 3.38VVADD785 pKa = 3.56VTVPDD790 pKa = 4.63PEE792 pKa = 4.74SPLLRR797 pKa = 11.84AVWSDD802 pKa = 2.72SHH804 pKa = 7.17GFVPPILALEE814 pKa = 4.44APVDD818 pKa = 3.89VDD820 pKa = 4.51GVLPPSGPARR830 pKa = 11.84TNGQRR835 pKa = 11.84LSRR838 pKa = 11.84VLQLPFGASDD848 pKa = 4.01LGRR851 pKa = 11.84ARR853 pKa = 11.84CSPPKK858 pKa = 10.18AEE860 pKa = 4.33SSGTPGLGVNVVPTPRR876 pKa = 11.84RR877 pKa = 11.84EE878 pKa = 4.04LRR880 pKa = 11.84NRR882 pKa = 11.84DD883 pKa = 3.52PLGLLGLNRR892 pKa = 11.84VGGQQAQQPPLNPPPPPAGADD913 pKa = 3.22PALANRR919 pKa = 11.84GAGQPAGAGNDD930 pKa = 3.86DD931 pKa = 3.44QHH933 pKa = 8.56DD934 pKa = 4.02PDD936 pKa = 4.13SRR938 pKa = 3.95
Molecular weight: 100.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFA5|A0A1L3KFA5_9VIRU RdRp catalytic domain-containing protein OS=Hubei toti-like virus 24 OX=1923313 PE=4 SV=1
MM1 pKa = 7.39MISTIRR7 pKa = 11.84TLGRR11 pKa = 11.84WAGPRR16 pKa = 11.84PTAKK20 pKa = 10.17LGHH23 pKa = 6.36FGGGRR28 pKa = 11.84VDD30 pKa = 4.97GKK32 pKa = 9.74LTLDD36 pKa = 3.63WAWVSGFEE44 pKa = 4.25FLPAGTALRR53 pKa = 11.84TLLKK57 pKa = 10.65GCDD60 pKa = 3.26VARR63 pKa = 11.84NFARR67 pKa = 11.84LEE69 pKa = 3.71GLRR72 pKa = 11.84RR73 pKa = 11.84SRR75 pKa = 11.84YY76 pKa = 7.51TSVKK80 pKa = 8.87HH81 pKa = 4.68TQQLVTWAGSFAGPLAFLLDD101 pKa = 3.74MSEE104 pKa = 4.22AAAAADD110 pKa = 3.33PVVPFFYY117 pKa = 10.67HH118 pKa = 7.17LFVTTAKK125 pKa = 10.5ARR127 pKa = 11.84VEE129 pKa = 4.07MAVEE133 pKa = 4.15VLSPDD138 pKa = 3.63STVSWPGVPKK148 pKa = 10.92LPDD151 pKa = 3.85HH152 pKa = 6.87PDD154 pKa = 3.18EE155 pKa = 4.39YY156 pKa = 11.04RR157 pKa = 11.84DD158 pKa = 4.05DD159 pKa = 3.71IVRR162 pKa = 11.84MVIKK166 pKa = 10.63DD167 pKa = 3.43MEE169 pKa = 4.53EE170 pKa = 4.06EE171 pKa = 4.36VSSHH175 pKa = 6.28GVDD178 pKa = 3.03ARR180 pKa = 11.84LAISGLYY187 pKa = 9.24TSGGPRR193 pKa = 11.84GWDD196 pKa = 3.2RR197 pKa = 11.84EE198 pKa = 4.19HH199 pKa = 6.24MVDD202 pKa = 5.1YY203 pKa = 10.41VKK205 pKa = 10.84SWIQGEE211 pKa = 4.31APLFSARR218 pKa = 11.84EE219 pKa = 3.66EE220 pKa = 4.01WVRR223 pKa = 11.84AKK225 pKa = 10.76LRR227 pKa = 11.84FWFSSWVKK235 pKa = 10.21LAPPDD240 pKa = 3.67GAGGFDD246 pKa = 3.47FEE248 pKa = 4.51QYY250 pKa = 10.45KK251 pKa = 10.36RR252 pKa = 11.84DD253 pKa = 3.65VIRR256 pKa = 11.84WGTSGGASPPPEE268 pKa = 3.52VRR270 pKa = 11.84EE271 pKa = 3.93RR272 pKa = 11.84LEE274 pKa = 4.24SLFPDD279 pKa = 4.02LPKK282 pKa = 10.64SAFRR286 pKa = 11.84TKK288 pKa = 7.8TTAGISSLLFGEE300 pKa = 5.17LEE302 pKa = 5.31DD303 pKa = 4.61FFKK306 pKa = 11.1PNNVCHH312 pKa = 6.59AALKK316 pKa = 9.72EE317 pKa = 3.92XAKK320 pKa = 9.98TRR322 pKa = 11.84IIVSTPMHH330 pKa = 6.84SYY332 pKa = 10.88VAMCFVFDD340 pKa = 5.2LLGPPHH346 pKa = 6.5FLKK349 pKa = 10.02STIASSRR356 pKa = 11.84GLEE359 pKa = 3.66EE360 pKa = 3.68FARR363 pKa = 11.84YY364 pKa = 7.78RR365 pKa = 11.84AKK367 pKa = 10.78YY368 pKa = 8.55YY369 pKa = 10.96AAIDD373 pKa = 3.6ASKK376 pKa = 10.47FDD378 pKa = 4.21HH379 pKa = 6.79NVPRR383 pKa = 11.84WLLEE387 pKa = 4.21MIWDD391 pKa = 4.01EE392 pKa = 4.34LALALEE398 pKa = 4.51SRR400 pKa = 11.84CDD402 pKa = 3.61EE403 pKa = 4.32RR404 pKa = 11.84FTTASHH410 pKa = 7.15LCRR413 pKa = 11.84SLSSEE418 pKa = 3.69LRR420 pKa = 11.84NLVVEE425 pKa = 4.12ILGTHH430 pKa = 6.02LRR432 pKa = 11.84YY433 pKa = 9.89EE434 pKa = 4.33KK435 pKa = 10.91GLLSGWRR442 pKa = 11.84ATSFIGSLVSALCCEE457 pKa = 4.98SYY459 pKa = 11.01VEE461 pKa = 4.24HH462 pKa = 6.81HH463 pKa = 6.71ASASRR468 pKa = 11.84SEE470 pKa = 3.82YY471 pKa = 10.43RR472 pKa = 11.84HH473 pKa = 5.91WIDD476 pKa = 4.44YY477 pKa = 9.67LVQGDD482 pKa = 4.72DD483 pKa = 3.86IILFSSAYY491 pKa = 9.83DD492 pKa = 3.58LSDD495 pKa = 3.04SVDD498 pKa = 3.34YY499 pKa = 10.39MRR501 pKa = 11.84GLGVLTHH508 pKa = 6.85PSKK511 pKa = 11.07CLIGADD517 pKa = 3.84GDD519 pKa = 3.91FLRR522 pKa = 11.84AIYY525 pKa = 9.68TKK527 pKa = 10.22DD528 pKa = 3.12GRR530 pKa = 11.84KK531 pKa = 7.33TYY533 pKa = 10.39PMRR536 pKa = 11.84ALRR539 pKa = 11.84SIFYY543 pKa = 10.22ANPWVEE549 pKa = 3.72QRR551 pKa = 11.84AFKK554 pKa = 10.8GVVEE558 pKa = 5.11ISQNWLVVEE567 pKa = 4.5SRR569 pKa = 11.84FSLLCTTSDD578 pKa = 3.0VEE580 pKa = 4.14RR581 pKa = 11.84AVRR584 pKa = 11.84SAACADD590 pKa = 3.27IARR593 pKa = 11.84WSGGSLRR600 pKa = 11.84ASQVRR605 pKa = 11.84ALYY608 pKa = 7.55NTPLSVGGLGPVEE621 pKa = 4.55SVFGTTVTTLTVDD634 pKa = 3.28YY635 pKa = 10.76DD636 pKa = 3.97VYY638 pKa = 11.37AEE640 pKa = 4.2KK641 pKa = 10.07TRR643 pKa = 11.84SVLPSLWQFVGVTVPLKK660 pKa = 8.78PSKK663 pKa = 10.51RR664 pKa = 11.84FTLQSVDD671 pKa = 2.99IFSLRR676 pKa = 11.84NLAKK680 pKa = 10.3KK681 pKa = 10.55AFFTSVPHH689 pKa = 7.14DD690 pKa = 4.03LDD692 pKa = 3.7KK693 pKa = 11.55KK694 pKa = 10.48GVNLTSLFLSVIYY707 pKa = 8.76DD708 pKa = 3.69TEE710 pKa = 4.2ARR712 pKa = 11.84RR713 pKa = 11.84HH714 pKa = 4.76TVDD717 pKa = 2.97AGSFEE722 pKa = 4.52VVALRR727 pKa = 11.84RR728 pKa = 11.84ALSIVGYY735 pKa = 8.39TDD737 pKa = 3.0YY738 pKa = 11.24RR739 pKa = 11.84PFVGGIYY746 pKa = 10.13RR747 pKa = 11.84CTTRR751 pKa = 11.84ASEE754 pKa = 4.22VLSLLFSGKK763 pKa = 10.1DD764 pKa = 3.24PSVPPSVYY772 pKa = 10.88QMTSPVSRR780 pKa = 11.84AVKK783 pKa = 9.59WLASLLGRR791 pKa = 11.84WLRR794 pKa = 11.84YY795 pKa = 8.92KK796 pKa = 10.71ARR798 pKa = 11.84TVKK801 pKa = 10.32GVRR804 pKa = 11.84AWSAGYY810 pKa = 8.11TLLCSAITVSTVKK823 pKa = 9.94GTMM826 pKa = 3.24
MM1 pKa = 7.39MISTIRR7 pKa = 11.84TLGRR11 pKa = 11.84WAGPRR16 pKa = 11.84PTAKK20 pKa = 10.17LGHH23 pKa = 6.36FGGGRR28 pKa = 11.84VDD30 pKa = 4.97GKK32 pKa = 9.74LTLDD36 pKa = 3.63WAWVSGFEE44 pKa = 4.25FLPAGTALRR53 pKa = 11.84TLLKK57 pKa = 10.65GCDD60 pKa = 3.26VARR63 pKa = 11.84NFARR67 pKa = 11.84LEE69 pKa = 3.71GLRR72 pKa = 11.84RR73 pKa = 11.84SRR75 pKa = 11.84YY76 pKa = 7.51TSVKK80 pKa = 8.87HH81 pKa = 4.68TQQLVTWAGSFAGPLAFLLDD101 pKa = 3.74MSEE104 pKa = 4.22AAAAADD110 pKa = 3.33PVVPFFYY117 pKa = 10.67HH118 pKa = 7.17LFVTTAKK125 pKa = 10.5ARR127 pKa = 11.84VEE129 pKa = 4.07MAVEE133 pKa = 4.15VLSPDD138 pKa = 3.63STVSWPGVPKK148 pKa = 10.92LPDD151 pKa = 3.85HH152 pKa = 6.87PDD154 pKa = 3.18EE155 pKa = 4.39YY156 pKa = 11.04RR157 pKa = 11.84DD158 pKa = 4.05DD159 pKa = 3.71IVRR162 pKa = 11.84MVIKK166 pKa = 10.63DD167 pKa = 3.43MEE169 pKa = 4.53EE170 pKa = 4.06EE171 pKa = 4.36VSSHH175 pKa = 6.28GVDD178 pKa = 3.03ARR180 pKa = 11.84LAISGLYY187 pKa = 9.24TSGGPRR193 pKa = 11.84GWDD196 pKa = 3.2RR197 pKa = 11.84EE198 pKa = 4.19HH199 pKa = 6.24MVDD202 pKa = 5.1YY203 pKa = 10.41VKK205 pKa = 10.84SWIQGEE211 pKa = 4.31APLFSARR218 pKa = 11.84EE219 pKa = 3.66EE220 pKa = 4.01WVRR223 pKa = 11.84AKK225 pKa = 10.76LRR227 pKa = 11.84FWFSSWVKK235 pKa = 10.21LAPPDD240 pKa = 3.67GAGGFDD246 pKa = 3.47FEE248 pKa = 4.51QYY250 pKa = 10.45KK251 pKa = 10.36RR252 pKa = 11.84DD253 pKa = 3.65VIRR256 pKa = 11.84WGTSGGASPPPEE268 pKa = 3.52VRR270 pKa = 11.84EE271 pKa = 3.93RR272 pKa = 11.84LEE274 pKa = 4.24SLFPDD279 pKa = 4.02LPKK282 pKa = 10.64SAFRR286 pKa = 11.84TKK288 pKa = 7.8TTAGISSLLFGEE300 pKa = 5.17LEE302 pKa = 5.31DD303 pKa = 4.61FFKK306 pKa = 11.1PNNVCHH312 pKa = 6.59AALKK316 pKa = 9.72EE317 pKa = 3.92XAKK320 pKa = 9.98TRR322 pKa = 11.84IIVSTPMHH330 pKa = 6.84SYY332 pKa = 10.88VAMCFVFDD340 pKa = 5.2LLGPPHH346 pKa = 6.5FLKK349 pKa = 10.02STIASSRR356 pKa = 11.84GLEE359 pKa = 3.66EE360 pKa = 3.68FARR363 pKa = 11.84YY364 pKa = 7.78RR365 pKa = 11.84AKK367 pKa = 10.78YY368 pKa = 8.55YY369 pKa = 10.96AAIDD373 pKa = 3.6ASKK376 pKa = 10.47FDD378 pKa = 4.21HH379 pKa = 6.79NVPRR383 pKa = 11.84WLLEE387 pKa = 4.21MIWDD391 pKa = 4.01EE392 pKa = 4.34LALALEE398 pKa = 4.51SRR400 pKa = 11.84CDD402 pKa = 3.61EE403 pKa = 4.32RR404 pKa = 11.84FTTASHH410 pKa = 7.15LCRR413 pKa = 11.84SLSSEE418 pKa = 3.69LRR420 pKa = 11.84NLVVEE425 pKa = 4.12ILGTHH430 pKa = 6.02LRR432 pKa = 11.84YY433 pKa = 9.89EE434 pKa = 4.33KK435 pKa = 10.91GLLSGWRR442 pKa = 11.84ATSFIGSLVSALCCEE457 pKa = 4.98SYY459 pKa = 11.01VEE461 pKa = 4.24HH462 pKa = 6.81HH463 pKa = 6.71ASASRR468 pKa = 11.84SEE470 pKa = 3.82YY471 pKa = 10.43RR472 pKa = 11.84HH473 pKa = 5.91WIDD476 pKa = 4.44YY477 pKa = 9.67LVQGDD482 pKa = 4.72DD483 pKa = 3.86IILFSSAYY491 pKa = 9.83DD492 pKa = 3.58LSDD495 pKa = 3.04SVDD498 pKa = 3.34YY499 pKa = 10.39MRR501 pKa = 11.84GLGVLTHH508 pKa = 6.85PSKK511 pKa = 11.07CLIGADD517 pKa = 3.84GDD519 pKa = 3.91FLRR522 pKa = 11.84AIYY525 pKa = 9.68TKK527 pKa = 10.22DD528 pKa = 3.12GRR530 pKa = 11.84KK531 pKa = 7.33TYY533 pKa = 10.39PMRR536 pKa = 11.84ALRR539 pKa = 11.84SIFYY543 pKa = 10.22ANPWVEE549 pKa = 3.72QRR551 pKa = 11.84AFKK554 pKa = 10.8GVVEE558 pKa = 5.11ISQNWLVVEE567 pKa = 4.5SRR569 pKa = 11.84FSLLCTTSDD578 pKa = 3.0VEE580 pKa = 4.14RR581 pKa = 11.84AVRR584 pKa = 11.84SAACADD590 pKa = 3.27IARR593 pKa = 11.84WSGGSLRR600 pKa = 11.84ASQVRR605 pKa = 11.84ALYY608 pKa = 7.55NTPLSVGGLGPVEE621 pKa = 4.55SVFGTTVTTLTVDD634 pKa = 3.28YY635 pKa = 10.76DD636 pKa = 3.97VYY638 pKa = 11.37AEE640 pKa = 4.2KK641 pKa = 10.07TRR643 pKa = 11.84SVLPSLWQFVGVTVPLKK660 pKa = 8.78PSKK663 pKa = 10.51RR664 pKa = 11.84FTLQSVDD671 pKa = 2.99IFSLRR676 pKa = 11.84NLAKK680 pKa = 10.3KK681 pKa = 10.55AFFTSVPHH689 pKa = 7.14DD690 pKa = 4.03LDD692 pKa = 3.7KK693 pKa = 11.55KK694 pKa = 10.48GVNLTSLFLSVIYY707 pKa = 8.76DD708 pKa = 3.69TEE710 pKa = 4.2ARR712 pKa = 11.84RR713 pKa = 11.84HH714 pKa = 4.76TVDD717 pKa = 2.97AGSFEE722 pKa = 4.52VVALRR727 pKa = 11.84RR728 pKa = 11.84ALSIVGYY735 pKa = 8.39TDD737 pKa = 3.0YY738 pKa = 11.24RR739 pKa = 11.84PFVGGIYY746 pKa = 10.13RR747 pKa = 11.84CTTRR751 pKa = 11.84ASEE754 pKa = 4.22VLSLLFSGKK763 pKa = 10.1DD764 pKa = 3.24PSVPPSVYY772 pKa = 10.88QMTSPVSRR780 pKa = 11.84AVKK783 pKa = 9.59WLASLLGRR791 pKa = 11.84WLRR794 pKa = 11.84YY795 pKa = 8.92KK796 pKa = 10.71ARR798 pKa = 11.84TVKK801 pKa = 10.32GVRR804 pKa = 11.84AWSAGYY810 pKa = 8.11TLLCSAITVSTVKK823 pKa = 9.94GTMM826 pKa = 3.24
Molecular weight: 92.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1764 |
826 |
938 |
882.0 |
96.44 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.921 ± 0.818 | 1.304 ± 0.101 |
5.556 ± 0.155 | 4.422 ± 0.368 |
4.082 ± 0.599 | 7.2 ± 0.121 |
1.984 ± 0.132 | 3.685 ± 0.2 |
3.571 ± 0.535 | 9.751 ± 0.367 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.814 ± 0.081 | 2.211 ± 0.679 |
6.122 ± 0.952 | 2.721 ± 0.944 |
6.689 ± 0.719 | 8.333 ± 0.836 |
6.463 ± 0.196 | 9.127 ± 0.361 |
2.324 ± 0.23 | 2.608 ± 0.613 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
