
Coconut foliar decay alphasatellite 7
Taxonomy: Viruses; Alphasatellitidae; Petromoalphasatellitinae; Coprasatellite
Average proteome isoelectric point is 5.57
Get precalculated fractions of proteins
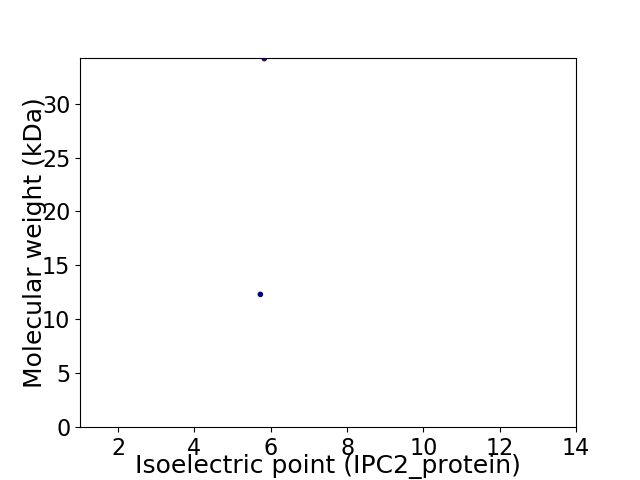
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2R4N9C1|A0A2R4N9C1_9VIRU Uncharacterized protein OS=Coconut foliar decay alphasatellite 7 OX=2161880 PE=4 SV=1
MM1 pKa = 8.51DD2 pKa = 4.1YY3 pKa = 11.08LKK5 pKa = 10.3PNNSISSVSLSSCTFYY21 pKa = 11.5KK22 pKa = 10.8LLVLQYY28 pKa = 10.74EE29 pKa = 4.67VLQHH33 pKa = 6.84LLRR36 pKa = 11.84TFHH39 pKa = 7.1KK40 pKa = 10.79AFVSNQDD47 pKa = 2.85ASEE50 pKa = 3.93LHH52 pKa = 5.66VRR54 pKa = 11.84KK55 pKa = 9.19EE56 pKa = 4.02HH57 pKa = 6.24FLSNNKK63 pKa = 10.11SYY65 pKa = 10.44FADD68 pKa = 3.82QIPSPVLNAQYY79 pKa = 10.47YY80 pKa = 10.58SLMSSSNLQAYY91 pKa = 9.81GPFLQDD97 pKa = 3.19EE98 pKa = 5.02EE99 pKa = 4.41ILAGAFVQSS108 pKa = 3.67
MM1 pKa = 8.51DD2 pKa = 4.1YY3 pKa = 11.08LKK5 pKa = 10.3PNNSISSVSLSSCTFYY21 pKa = 11.5KK22 pKa = 10.8LLVLQYY28 pKa = 10.74EE29 pKa = 4.67VLQHH33 pKa = 6.84LLRR36 pKa = 11.84TFHH39 pKa = 7.1KK40 pKa = 10.79AFVSNQDD47 pKa = 2.85ASEE50 pKa = 3.93LHH52 pKa = 5.66VRR54 pKa = 11.84KK55 pKa = 9.19EE56 pKa = 4.02HH57 pKa = 6.24FLSNNKK63 pKa = 10.11SYY65 pKa = 10.44FADD68 pKa = 3.82QIPSPVLNAQYY79 pKa = 10.47YY80 pKa = 10.58SLMSSSNLQAYY91 pKa = 9.81GPFLQDD97 pKa = 3.19EE98 pKa = 5.02EE99 pKa = 4.41ILAGAFVQSS108 pKa = 3.67
Molecular weight: 12.31 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2R4N9C1|A0A2R4N9C1_9VIRU Uncharacterized protein OS=Coconut foliar decay alphasatellite 7 OX=2161880 PE=4 SV=1
MM1 pKa = 7.64ASQVRR6 pKa = 11.84RR7 pKa = 11.84WVLTFNYY14 pKa = 9.27SDD16 pKa = 3.87EE17 pKa = 4.44SAASDD22 pKa = 3.34LVRR25 pKa = 11.84RR26 pKa = 11.84IEE28 pKa = 4.11SLKK31 pKa = 10.62LIYY34 pKa = 10.16GIIGDD39 pKa = 4.99EE40 pKa = 4.06IAPTTGQRR48 pKa = 11.84HH49 pKa = 4.74LQGFLHH55 pKa = 6.47LAGRR59 pKa = 11.84GRR61 pKa = 11.84TLEE64 pKa = 4.04GLKK67 pKa = 8.93KK68 pKa = 8.74TLEE71 pKa = 4.22NNTVHH76 pKa = 7.11LEE78 pKa = 3.9PAKK81 pKa = 10.96GSDD84 pKa = 3.59QQNKK88 pKa = 9.47IYY90 pKa = 10.28CSKK93 pKa = 10.69EE94 pKa = 3.58NVLFEE99 pKa = 4.83HH100 pKa = 7.03GVPTRR105 pKa = 11.84PGSKK109 pKa = 8.68RR110 pKa = 11.84KK111 pKa = 9.79LYY113 pKa = 10.34EE114 pKa = 4.04RR115 pKa = 11.84YY116 pKa = 10.07EE117 pKa = 4.06EE118 pKa = 4.29DD119 pKa = 3.68AEE121 pKa = 4.09EE122 pKa = 4.54LRR124 pKa = 11.84IEE126 pKa = 4.18EE127 pKa = 4.13PGAYY131 pKa = 9.04RR132 pKa = 11.84RR133 pKa = 11.84CKK135 pKa = 10.05MIEE138 pKa = 3.95KK139 pKa = 9.13QKK141 pKa = 10.57KK142 pKa = 6.14WNYY145 pKa = 8.45WVLNNPFPYY154 pKa = 10.28DD155 pKa = 3.75LYY157 pKa = 10.68QWQEE161 pKa = 3.65EE162 pKa = 4.27LMEE165 pKa = 4.3VLQEE169 pKa = 4.09PADD172 pKa = 3.59NRR174 pKa = 11.84TILWICGRR182 pKa = 11.84DD183 pKa = 3.5GGDD186 pKa = 2.94GKK188 pKa = 9.12TEE190 pKa = 3.9FGKK193 pKa = 11.05YY194 pKa = 9.9LGLNEE199 pKa = 4.03KK200 pKa = 9.22WFYY203 pKa = 11.15SCGGKK208 pKa = 7.83TRR210 pKa = 11.84DD211 pKa = 3.3VLYY214 pKa = 10.55QYY216 pKa = 10.6IEE218 pKa = 3.91EE219 pKa = 4.4PEE221 pKa = 4.12RR222 pKa = 11.84NFILDD227 pKa = 3.44VPRR230 pKa = 11.84CNDD233 pKa = 2.51QYY235 pKa = 11.41MNYY238 pKa = 11.05ALIEE242 pKa = 4.52CIKK245 pKa = 10.66NRR247 pKa = 11.84AFSSDD252 pKa = 3.0KK253 pKa = 11.21YY254 pKa = 11.25EE255 pKa = 4.06PLQYY259 pKa = 11.02LGFDD263 pKa = 3.33KK264 pKa = 11.23VHH266 pKa = 6.87VIVLCNFLPDD276 pKa = 3.64YY277 pKa = 11.15LKK279 pKa = 10.63ISEE282 pKa = 4.88DD283 pKa = 4.93RR284 pKa = 11.84IKK286 pKa = 10.8LWNII290 pKa = 3.27
MM1 pKa = 7.64ASQVRR6 pKa = 11.84RR7 pKa = 11.84WVLTFNYY14 pKa = 9.27SDD16 pKa = 3.87EE17 pKa = 4.44SAASDD22 pKa = 3.34LVRR25 pKa = 11.84RR26 pKa = 11.84IEE28 pKa = 4.11SLKK31 pKa = 10.62LIYY34 pKa = 10.16GIIGDD39 pKa = 4.99EE40 pKa = 4.06IAPTTGQRR48 pKa = 11.84HH49 pKa = 4.74LQGFLHH55 pKa = 6.47LAGRR59 pKa = 11.84GRR61 pKa = 11.84TLEE64 pKa = 4.04GLKK67 pKa = 8.93KK68 pKa = 8.74TLEE71 pKa = 4.22NNTVHH76 pKa = 7.11LEE78 pKa = 3.9PAKK81 pKa = 10.96GSDD84 pKa = 3.59QQNKK88 pKa = 9.47IYY90 pKa = 10.28CSKK93 pKa = 10.69EE94 pKa = 3.58NVLFEE99 pKa = 4.83HH100 pKa = 7.03GVPTRR105 pKa = 11.84PGSKK109 pKa = 8.68RR110 pKa = 11.84KK111 pKa = 9.79LYY113 pKa = 10.34EE114 pKa = 4.04RR115 pKa = 11.84YY116 pKa = 10.07EE117 pKa = 4.06EE118 pKa = 4.29DD119 pKa = 3.68AEE121 pKa = 4.09EE122 pKa = 4.54LRR124 pKa = 11.84IEE126 pKa = 4.18EE127 pKa = 4.13PGAYY131 pKa = 9.04RR132 pKa = 11.84RR133 pKa = 11.84CKK135 pKa = 10.05MIEE138 pKa = 3.95KK139 pKa = 9.13QKK141 pKa = 10.57KK142 pKa = 6.14WNYY145 pKa = 8.45WVLNNPFPYY154 pKa = 10.28DD155 pKa = 3.75LYY157 pKa = 10.68QWQEE161 pKa = 3.65EE162 pKa = 4.27LMEE165 pKa = 4.3VLQEE169 pKa = 4.09PADD172 pKa = 3.59NRR174 pKa = 11.84TILWICGRR182 pKa = 11.84DD183 pKa = 3.5GGDD186 pKa = 2.94GKK188 pKa = 9.12TEE190 pKa = 3.9FGKK193 pKa = 11.05YY194 pKa = 9.9LGLNEE199 pKa = 4.03KK200 pKa = 9.22WFYY203 pKa = 11.15SCGGKK208 pKa = 7.83TRR210 pKa = 11.84DD211 pKa = 3.3VLYY214 pKa = 10.55QYY216 pKa = 10.6IEE218 pKa = 3.91EE219 pKa = 4.4PEE221 pKa = 4.12RR222 pKa = 11.84NFILDD227 pKa = 3.44VPRR230 pKa = 11.84CNDD233 pKa = 2.51QYY235 pKa = 11.41MNYY238 pKa = 11.05ALIEE242 pKa = 4.52CIKK245 pKa = 10.66NRR247 pKa = 11.84AFSSDD252 pKa = 3.0KK253 pKa = 11.21YY254 pKa = 11.25EE255 pKa = 4.06PLQYY259 pKa = 11.02LGFDD263 pKa = 3.33KK264 pKa = 11.23VHH266 pKa = 6.87VIVLCNFLPDD276 pKa = 3.64YY277 pKa = 11.15LKK279 pKa = 10.63ISEE282 pKa = 4.88DD283 pKa = 4.93RR284 pKa = 11.84IKK286 pKa = 10.8LWNII290 pKa = 3.27
Molecular weight: 34.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
398 |
108 |
290 |
199.0 |
23.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.523 ± 0.968 | 2.01 ± 0.536 |
5.025 ± 0.653 | 8.291 ± 1.809 |
4.271 ± 1.092 | 5.528 ± 1.816 |
2.261 ± 0.713 | 5.276 ± 1.234 |
6.533 ± 0.94 | 11.307 ± 1.276 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.508 ± 0.17 | 5.779 ± 0.347 |
4.02 ± 0.156 | 5.025 ± 1.177 |
5.528 ± 1.816 | 7.035 ± 3.843 |
3.015 ± 0.575 | 5.025 ± 0.719 |
1.759 ± 0.869 | 6.281 ± 0.099 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
