
Cellulophaga algicola (strain DSM 14237 / IC166 / ACAM 630)
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Cellulophaga; Cellulophaga algicola
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
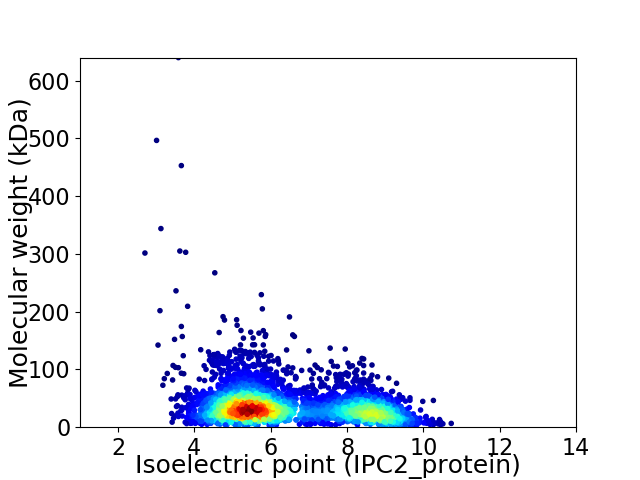
Virtual 2D-PAGE plot for 4114 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E6X867|E6X867_CELAD Transcriptional regulator AraC family OS=Cellulophaga algicola (strain DSM 14237 / IC166 / ACAM 630) OX=688270 GN=Celal_2403 PE=4 SV=1
MM1 pKa = 7.39SVTPTTEE8 pKa = 4.58CLDD11 pKa = 3.67SDD13 pKa = 4.08NDD15 pKa = 4.02GVPDD19 pKa = 4.58VFDD22 pKa = 5.52LDD24 pKa = 3.82SDD26 pKa = 3.77NDD28 pKa = 4.58GIYY31 pKa = 10.79DD32 pKa = 3.65AVEE35 pKa = 4.13AGHH38 pKa = 6.05NQAHH42 pKa = 6.35TDD44 pKa = 3.73GVVTGAVGTDD54 pKa = 3.48GVPDD58 pKa = 3.68NVQNDD63 pKa = 3.99PNRR66 pKa = 11.84EE67 pKa = 4.03TVNYY71 pKa = 8.24TLSDD75 pKa = 3.75SDD77 pKa = 5.46LDD79 pKa = 4.2TIPDD83 pKa = 3.6VLEE86 pKa = 4.3FDD88 pKa = 5.13SDD90 pKa = 3.78NDD92 pKa = 3.76GCNDD96 pKa = 3.16SDD98 pKa = 3.67EE99 pKa = 5.36AYY101 pKa = 10.18GAKK104 pKa = 9.48DD105 pKa = 3.22TDD107 pKa = 3.43SDD109 pKa = 4.23ANGFYY114 pKa = 10.91GSGQPNVDD122 pKa = 3.13VNGRR126 pKa = 11.84ITAATYY132 pKa = 9.26PEE134 pKa = 4.93PNDD137 pKa = 3.58GDD139 pKa = 4.38SNTVYY144 pKa = 10.54DD145 pKa = 4.2YY146 pKa = 11.42KK147 pKa = 10.81EE148 pKa = 4.17KK149 pKa = 10.67KK150 pKa = 8.43QAPIIADD157 pKa = 3.63KK158 pKa = 11.55NNTTIQACYY167 pKa = 8.85STDD170 pKa = 3.03VTLINSALYY179 pKa = 10.66ADD181 pKa = 4.42TFQWQLLNGSNWIDD195 pKa = 3.08ISDD198 pKa = 3.69STKK201 pKa = 10.68YY202 pKa = 10.68SGTGTNTLDD211 pKa = 4.0IINVTLTEE219 pKa = 4.18NGNQYY224 pKa = 11.1RR225 pKa = 11.84LIASHH230 pKa = 6.88SSTICDD236 pKa = 3.47EE237 pKa = 4.68DD238 pKa = 4.43SSGVTTLNVNDD249 pKa = 4.32EE250 pKa = 3.86MDD252 pKa = 3.9APVSGGDD259 pKa = 3.28QSYY262 pKa = 11.17CSGDD266 pKa = 3.98SIPQLSANVPSDD278 pKa = 3.62EE279 pKa = 4.35TVDD282 pKa = 3.23WYY284 pKa = 11.25ANLSGGTALLEE295 pKa = 4.41SSLSYY300 pKa = 10.54TPAGAGTYY308 pKa = 8.72YY309 pKa = 11.1AEE311 pKa = 4.42ARR313 pKa = 11.84STTFVGCTSTTRR325 pKa = 11.84TPITLTEE332 pKa = 4.04EE333 pKa = 4.26SPSVVTIGADD343 pKa = 2.98QVVFVGDD350 pKa = 3.54NAIFTATASNSDD362 pKa = 3.52TFHH365 pKa = 6.86WEE367 pKa = 3.45VSTDD371 pKa = 3.16GGITFNSVAEE381 pKa = 4.14SSEE384 pKa = 4.02YY385 pKa = 10.65TGTQTVTLTVVSARR399 pKa = 11.84ALQNGYY405 pKa = 9.09RR406 pKa = 11.84FRR408 pKa = 11.84FVASTAGSSCGTTNSSSAVLTVKK431 pKa = 10.52VKK433 pKa = 9.52TVITNRR439 pKa = 11.84RR440 pKa = 11.84ITYY443 pKa = 9.56RR444 pKa = 11.84VKK446 pKa = 10.88KK447 pKa = 10.21NN448 pKa = 3.02
MM1 pKa = 7.39SVTPTTEE8 pKa = 4.58CLDD11 pKa = 3.67SDD13 pKa = 4.08NDD15 pKa = 4.02GVPDD19 pKa = 4.58VFDD22 pKa = 5.52LDD24 pKa = 3.82SDD26 pKa = 3.77NDD28 pKa = 4.58GIYY31 pKa = 10.79DD32 pKa = 3.65AVEE35 pKa = 4.13AGHH38 pKa = 6.05NQAHH42 pKa = 6.35TDD44 pKa = 3.73GVVTGAVGTDD54 pKa = 3.48GVPDD58 pKa = 3.68NVQNDD63 pKa = 3.99PNRR66 pKa = 11.84EE67 pKa = 4.03TVNYY71 pKa = 8.24TLSDD75 pKa = 3.75SDD77 pKa = 5.46LDD79 pKa = 4.2TIPDD83 pKa = 3.6VLEE86 pKa = 4.3FDD88 pKa = 5.13SDD90 pKa = 3.78NDD92 pKa = 3.76GCNDD96 pKa = 3.16SDD98 pKa = 3.67EE99 pKa = 5.36AYY101 pKa = 10.18GAKK104 pKa = 9.48DD105 pKa = 3.22TDD107 pKa = 3.43SDD109 pKa = 4.23ANGFYY114 pKa = 10.91GSGQPNVDD122 pKa = 3.13VNGRR126 pKa = 11.84ITAATYY132 pKa = 9.26PEE134 pKa = 4.93PNDD137 pKa = 3.58GDD139 pKa = 4.38SNTVYY144 pKa = 10.54DD145 pKa = 4.2YY146 pKa = 11.42KK147 pKa = 10.81EE148 pKa = 4.17KK149 pKa = 10.67KK150 pKa = 8.43QAPIIADD157 pKa = 3.63KK158 pKa = 11.55NNTTIQACYY167 pKa = 8.85STDD170 pKa = 3.03VTLINSALYY179 pKa = 10.66ADD181 pKa = 4.42TFQWQLLNGSNWIDD195 pKa = 3.08ISDD198 pKa = 3.69STKK201 pKa = 10.68YY202 pKa = 10.68SGTGTNTLDD211 pKa = 4.0IINVTLTEE219 pKa = 4.18NGNQYY224 pKa = 11.1RR225 pKa = 11.84LIASHH230 pKa = 6.88SSTICDD236 pKa = 3.47EE237 pKa = 4.68DD238 pKa = 4.43SSGVTTLNVNDD249 pKa = 4.32EE250 pKa = 3.86MDD252 pKa = 3.9APVSGGDD259 pKa = 3.28QSYY262 pKa = 11.17CSGDD266 pKa = 3.98SIPQLSANVPSDD278 pKa = 3.62EE279 pKa = 4.35TVDD282 pKa = 3.23WYY284 pKa = 11.25ANLSGGTALLEE295 pKa = 4.41SSLSYY300 pKa = 10.54TPAGAGTYY308 pKa = 8.72YY309 pKa = 11.1AEE311 pKa = 4.42ARR313 pKa = 11.84STTFVGCTSTTRR325 pKa = 11.84TPITLTEE332 pKa = 4.04EE333 pKa = 4.26SPSVVTIGADD343 pKa = 2.98QVVFVGDD350 pKa = 3.54NAIFTATASNSDD362 pKa = 3.52TFHH365 pKa = 6.86WEE367 pKa = 3.45VSTDD371 pKa = 3.16GGITFNSVAEE381 pKa = 4.14SSEE384 pKa = 4.02YY385 pKa = 10.65TGTQTVTLTVVSARR399 pKa = 11.84ALQNGYY405 pKa = 9.09RR406 pKa = 11.84FRR408 pKa = 11.84FVASTAGSSCGTTNSSSAVLTVKK431 pKa = 10.52VKK433 pKa = 9.52TVITNRR439 pKa = 11.84RR440 pKa = 11.84ITYY443 pKa = 9.56RR444 pKa = 11.84VKK446 pKa = 10.88KK447 pKa = 10.21NN448 pKa = 3.02
Molecular weight: 47.55 kDa
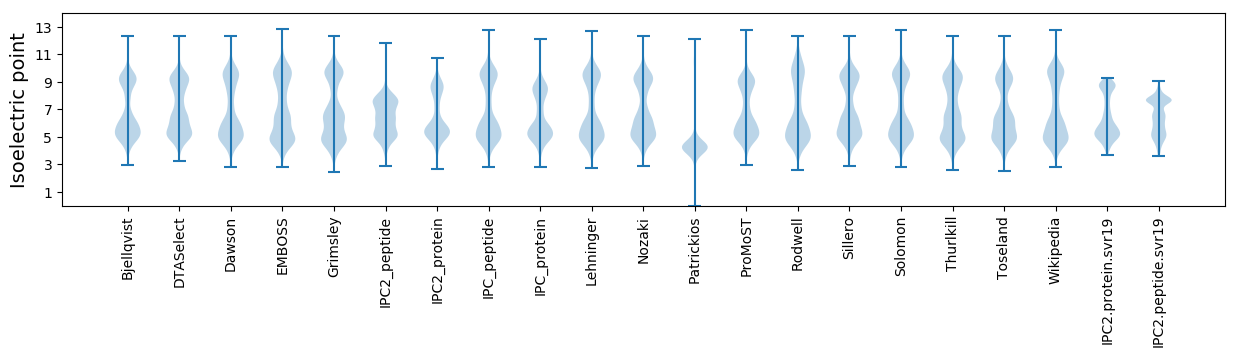
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E6XCM7|E6XCM7_CELAD Glutamate 5-kinase OS=Cellulophaga algicola (strain DSM 14237 / IC166 / ACAM 630) OX=688270 GN=proB PE=3 SV=1
MM1 pKa = 7.5AKK3 pKa = 10.25EE4 pKa = 3.96SMKK7 pKa = 10.52ARR9 pKa = 11.84EE10 pKa = 4.29VKK12 pKa = 10.32RR13 pKa = 11.84EE14 pKa = 3.88KK15 pKa = 10.1TVAKK19 pKa = 10.55YY20 pKa = 10.61ADD22 pKa = 3.25KK23 pKa = 11.03RR24 pKa = 11.84KK25 pKa = 9.78ALKK28 pKa = 10.38AAGDD32 pKa = 3.87YY33 pKa = 10.79EE34 pKa = 4.59GLQKK38 pKa = 10.72LPRR41 pKa = 11.84NANPIRR47 pKa = 11.84LHH49 pKa = 5.75NRR51 pKa = 11.84CKK53 pKa = 10.01LTGRR57 pKa = 11.84PRR59 pKa = 11.84GYY61 pKa = 9.14MRR63 pKa = 11.84TFGVSRR69 pKa = 11.84VTFRR73 pKa = 11.84EE74 pKa = 3.87MANEE78 pKa = 3.69GLIPGVRR85 pKa = 11.84KK86 pKa = 10.15ASWW89 pKa = 2.78
MM1 pKa = 7.5AKK3 pKa = 10.25EE4 pKa = 3.96SMKK7 pKa = 10.52ARR9 pKa = 11.84EE10 pKa = 4.29VKK12 pKa = 10.32RR13 pKa = 11.84EE14 pKa = 3.88KK15 pKa = 10.1TVAKK19 pKa = 10.55YY20 pKa = 10.61ADD22 pKa = 3.25KK23 pKa = 11.03RR24 pKa = 11.84KK25 pKa = 9.78ALKK28 pKa = 10.38AAGDD32 pKa = 3.87YY33 pKa = 10.79EE34 pKa = 4.59GLQKK38 pKa = 10.72LPRR41 pKa = 11.84NANPIRR47 pKa = 11.84LHH49 pKa = 5.75NRR51 pKa = 11.84CKK53 pKa = 10.01LTGRR57 pKa = 11.84PRR59 pKa = 11.84GYY61 pKa = 9.14MRR63 pKa = 11.84TFGVSRR69 pKa = 11.84VTFRR73 pKa = 11.84EE74 pKa = 3.87MANEE78 pKa = 3.69GLIPGVRR85 pKa = 11.84KK86 pKa = 10.15ASWW89 pKa = 2.78
Molecular weight: 10.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1411819 |
30 |
6081 |
343.2 |
38.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.583 ± 0.043 | 0.736 ± 0.011 |
5.729 ± 0.059 | 6.635 ± 0.046 |
5.184 ± 0.033 | 6.418 ± 0.05 |
1.676 ± 0.02 | 8.053 ± 0.036 |
7.769 ± 0.067 | 9.359 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.086 ± 0.021 | 6.085 ± 0.042 |
3.296 ± 0.027 | 3.154 ± 0.023 |
3.14 ± 0.028 | 6.595 ± 0.035 |
6.224 ± 0.071 | 6.127 ± 0.037 |
1.045 ± 0.015 | 4.105 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
