
Litorimicrobium taeanense
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Litorimicrobium
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
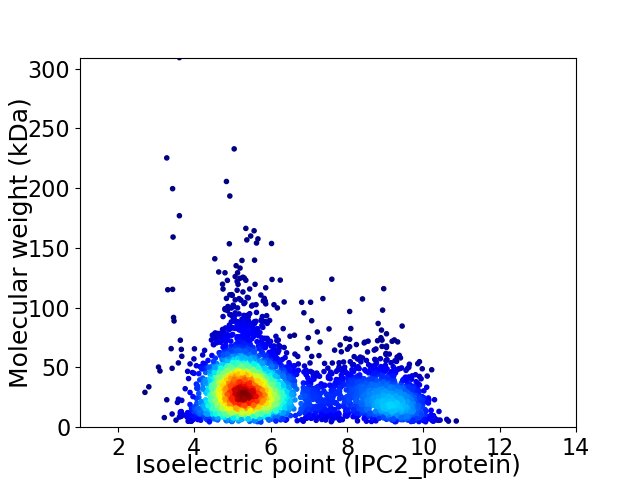
Virtual 2D-PAGE plot for 3839 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H9FAY9|A0A1H9FAY9_9RHOB Uncharacterized protein OS=Litorimicrobium taeanense OX=657014 GN=SAMN04488092_10617 PE=4 SV=1
MM1 pKa = 7.53AVKK4 pKa = 10.29QIVTGTTADD13 pKa = 3.47EE14 pKa = 4.27TLEE17 pKa = 4.11LTSGWGRR24 pKa = 11.84ISAGGGDD31 pKa = 3.81DD32 pKa = 3.57TLYY35 pKa = 10.35ATAQNGGNGTQGHH48 pKa = 5.55EE49 pKa = 3.84VHH51 pKa = 6.83TYY53 pKa = 10.07GGSGDD58 pKa = 4.02DD59 pKa = 3.47VTYY62 pKa = 11.4LDD64 pKa = 4.66FNSITSFSAGHH75 pKa = 6.51HH76 pKa = 5.78ARR78 pKa = 11.84GGTNDD83 pKa = 3.1GMSFSEE89 pKa = 4.28QNSMPSTDD97 pKa = 3.78HH98 pKa = 6.94IIPVSAHH105 pKa = 5.78SGSNVVSEE113 pKa = 4.51GVDD116 pKa = 3.04TFVFTNLQDD125 pKa = 3.34VGGLVVGRR133 pKa = 11.84LEE135 pKa = 4.37DD136 pKa = 4.1LEE138 pKa = 4.42HH139 pKa = 7.11SRR141 pKa = 11.84DD142 pKa = 3.56SIYY145 pKa = 10.74INSIGSNNEE154 pKa = 3.26INLSNNSGTVNGNSWRR170 pKa = 11.84IVEE173 pKa = 4.2WNGNHH178 pKa = 6.78IDD180 pKa = 4.13GNADD184 pKa = 3.27PQQWLMIQTTGGGTLLYY201 pKa = 9.7ALEE204 pKa = 4.42GARR207 pKa = 11.84VDD209 pKa = 5.36DD210 pKa = 4.84SPLTPGGEE218 pKa = 4.27QEE220 pKa = 4.3SHH222 pKa = 5.19FVSMNSATSFVNSNPVSVSFIDD244 pKa = 3.73QIDD247 pKa = 4.34IIPVGFSPDD256 pKa = 3.28AGGLLLDD263 pKa = 5.68DD264 pKa = 5.06YY265 pKa = 11.01DD266 pKa = 4.38TEE268 pKa = 4.33QSDD271 pKa = 3.79VDD273 pKa = 3.92SSINGTQEE281 pKa = 3.48GDD283 pKa = 3.65VIAAGLNDD291 pKa = 4.91DD292 pKa = 4.55SVTANGGDD300 pKa = 3.5DD301 pKa = 4.29TIWGGSGNDD310 pKa = 3.72VIDD313 pKa = 4.52GGSGDD318 pKa = 4.5DD319 pKa = 4.13SLDD322 pKa = 3.76GGSGADD328 pKa = 3.91TIIGGAGNDD337 pKa = 3.86VISGGSASSDD347 pKa = 3.27PRR349 pKa = 11.84DD350 pKa = 3.59VIYY353 pKa = 10.66GGNGNDD359 pKa = 4.42YY360 pKa = 11.0IDD362 pKa = 3.93GGYY365 pKa = 11.05GNDD368 pKa = 3.9EE369 pKa = 4.29LRR371 pKa = 11.84GDD373 pKa = 4.73DD374 pKa = 4.49GNDD377 pKa = 3.47TITGGFGADD386 pKa = 3.47SVIGGAGDD394 pKa = 4.66DD395 pKa = 4.0EE396 pKa = 6.06LNGQSLSDD404 pKa = 4.18EE405 pKa = 4.27IFGGDD410 pKa = 3.75GEE412 pKa = 5.05DD413 pKa = 4.58FINGGFGHH421 pKa = 7.23DD422 pKa = 3.98RR423 pKa = 11.84LNGGADD429 pKa = 3.35ADD431 pKa = 4.3RR432 pKa = 11.84FFHH435 pKa = 7.21LGIADD440 pKa = 4.56HH441 pKa = 6.66GSDD444 pKa = 3.81WIQDD448 pKa = 3.35YY449 pKa = 11.0SAAQGDD455 pKa = 3.98VLQYY459 pKa = 11.29GGTASANQFQVNFAATSGSGEE480 pKa = 4.11EE481 pKa = 4.63GVQEE485 pKa = 4.4AFVIYY490 pKa = 10.41QPTGQILWVLIDD502 pKa = 3.41GHH504 pKa = 7.02AQDD507 pKa = 5.46EE508 pKa = 4.72INLLLGGTQHH518 pKa = 7.58DD519 pKa = 4.49LLAA522 pKa = 5.63
MM1 pKa = 7.53AVKK4 pKa = 10.29QIVTGTTADD13 pKa = 3.47EE14 pKa = 4.27TLEE17 pKa = 4.11LTSGWGRR24 pKa = 11.84ISAGGGDD31 pKa = 3.81DD32 pKa = 3.57TLYY35 pKa = 10.35ATAQNGGNGTQGHH48 pKa = 5.55EE49 pKa = 3.84VHH51 pKa = 6.83TYY53 pKa = 10.07GGSGDD58 pKa = 4.02DD59 pKa = 3.47VTYY62 pKa = 11.4LDD64 pKa = 4.66FNSITSFSAGHH75 pKa = 6.51HH76 pKa = 5.78ARR78 pKa = 11.84GGTNDD83 pKa = 3.1GMSFSEE89 pKa = 4.28QNSMPSTDD97 pKa = 3.78HH98 pKa = 6.94IIPVSAHH105 pKa = 5.78SGSNVVSEE113 pKa = 4.51GVDD116 pKa = 3.04TFVFTNLQDD125 pKa = 3.34VGGLVVGRR133 pKa = 11.84LEE135 pKa = 4.37DD136 pKa = 4.1LEE138 pKa = 4.42HH139 pKa = 7.11SRR141 pKa = 11.84DD142 pKa = 3.56SIYY145 pKa = 10.74INSIGSNNEE154 pKa = 3.26INLSNNSGTVNGNSWRR170 pKa = 11.84IVEE173 pKa = 4.2WNGNHH178 pKa = 6.78IDD180 pKa = 4.13GNADD184 pKa = 3.27PQQWLMIQTTGGGTLLYY201 pKa = 9.7ALEE204 pKa = 4.42GARR207 pKa = 11.84VDD209 pKa = 5.36DD210 pKa = 4.84SPLTPGGEE218 pKa = 4.27QEE220 pKa = 4.3SHH222 pKa = 5.19FVSMNSATSFVNSNPVSVSFIDD244 pKa = 3.73QIDD247 pKa = 4.34IIPVGFSPDD256 pKa = 3.28AGGLLLDD263 pKa = 5.68DD264 pKa = 5.06YY265 pKa = 11.01DD266 pKa = 4.38TEE268 pKa = 4.33QSDD271 pKa = 3.79VDD273 pKa = 3.92SSINGTQEE281 pKa = 3.48GDD283 pKa = 3.65VIAAGLNDD291 pKa = 4.91DD292 pKa = 4.55SVTANGGDD300 pKa = 3.5DD301 pKa = 4.29TIWGGSGNDD310 pKa = 3.72VIDD313 pKa = 4.52GGSGDD318 pKa = 4.5DD319 pKa = 4.13SLDD322 pKa = 3.76GGSGADD328 pKa = 3.91TIIGGAGNDD337 pKa = 3.86VISGGSASSDD347 pKa = 3.27PRR349 pKa = 11.84DD350 pKa = 3.59VIYY353 pKa = 10.66GGNGNDD359 pKa = 4.42YY360 pKa = 11.0IDD362 pKa = 3.93GGYY365 pKa = 11.05GNDD368 pKa = 3.9EE369 pKa = 4.29LRR371 pKa = 11.84GDD373 pKa = 4.73DD374 pKa = 4.49GNDD377 pKa = 3.47TITGGFGADD386 pKa = 3.47SVIGGAGDD394 pKa = 4.66DD395 pKa = 4.0EE396 pKa = 6.06LNGQSLSDD404 pKa = 4.18EE405 pKa = 4.27IFGGDD410 pKa = 3.75GEE412 pKa = 5.05DD413 pKa = 4.58FINGGFGHH421 pKa = 7.23DD422 pKa = 3.98RR423 pKa = 11.84LNGGADD429 pKa = 3.35ADD431 pKa = 4.3RR432 pKa = 11.84FFHH435 pKa = 7.21LGIADD440 pKa = 4.56HH441 pKa = 6.66GSDD444 pKa = 3.81WIQDD448 pKa = 3.35YY449 pKa = 11.0SAAQGDD455 pKa = 3.98VLQYY459 pKa = 11.29GGTASANQFQVNFAATSGSGEE480 pKa = 4.11EE481 pKa = 4.63GVQEE485 pKa = 4.4AFVIYY490 pKa = 10.41QPTGQILWVLIDD502 pKa = 3.41GHH504 pKa = 7.02AQDD507 pKa = 5.46EE508 pKa = 4.72INLLLGGTQHH518 pKa = 7.58DD519 pKa = 4.49LLAA522 pKa = 5.63
Molecular weight: 53.79 kDa
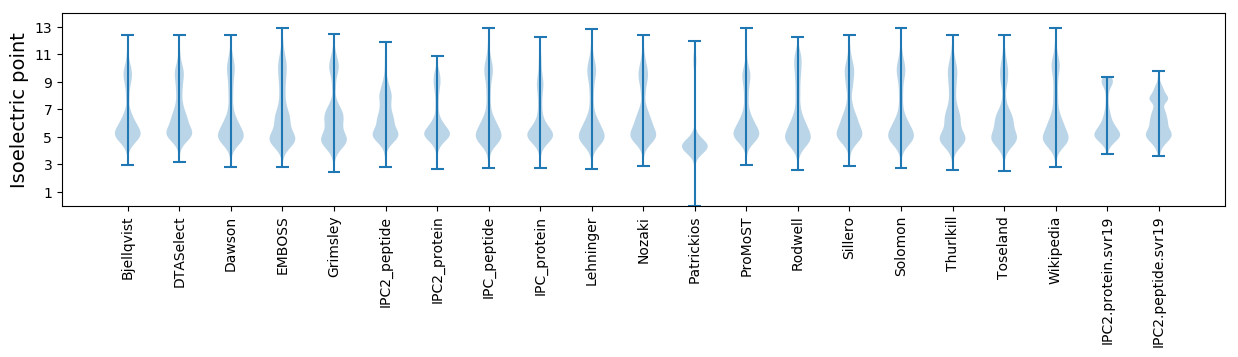
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H9IVY2|A0A1H9IVY2_9RHOB Uncharacterized protein OS=Litorimicrobium taeanense OX=657014 GN=SAMN04488092_11348 PE=4 SV=1
MM1 pKa = 7.06TCAKK5 pKa = 9.92VRR7 pKa = 11.84LKK9 pKa = 11.08APLRR13 pKa = 11.84RR14 pKa = 11.84GLTRR18 pKa = 11.84SHH20 pKa = 5.76AHH22 pKa = 6.57KK23 pKa = 10.57SHH25 pKa = 6.07NQDD28 pKa = 2.39HH29 pKa = 5.57VVVVQFWYY37 pKa = 10.17AYY39 pKa = 10.11LPEE42 pKa = 5.24GSSKK46 pKa = 10.46MSWQKK51 pKa = 10.31ISRR54 pKa = 11.84PTVPNQRR61 pKa = 11.84GRR63 pKa = 11.84EE64 pKa = 3.72RR65 pKa = 11.84RR66 pKa = 11.84DD67 pKa = 3.43CIKK70 pKa = 10.38IGSLAAFQNSRR81 pKa = 11.84TISYY85 pKa = 8.53GVPLDD90 pKa = 3.44KK91 pKa = 10.7RR92 pKa = 11.84KK93 pKa = 9.63QRR95 pKa = 11.84AVSSLCLGAAPVDD108 pKa = 3.39QLNCHH113 pKa = 5.93RR114 pKa = 11.84HH115 pKa = 4.44LHH117 pKa = 5.61SRR119 pKa = 11.84PLPRR123 pKa = 11.84RR124 pKa = 11.84NRR126 pKa = 11.84MEE128 pKa = 4.59RR129 pKa = 11.84GPDD132 pKa = 3.19QKK134 pKa = 10.7STLNRR139 pKa = 11.84IRR141 pKa = 11.84LNLKK145 pKa = 8.7SVRR148 pKa = 11.84TCATLASGRR157 pKa = 11.84PIPNLLSPLITLFCADD173 pKa = 4.47HH174 pKa = 6.74SDD176 pKa = 3.94CEE178 pKa = 4.3KK179 pKa = 11.01ASTRR183 pKa = 11.84TEE185 pKa = 3.87RR186 pKa = 11.84LLPDD190 pKa = 3.68PTAPHH195 pKa = 7.49PSRR198 pKa = 11.84FHH200 pKa = 6.7PKK202 pKa = 9.76RR203 pKa = 11.84EE204 pKa = 4.0QVSDD208 pKa = 4.0DD209 pKa = 3.54SATARR214 pKa = 11.84DD215 pKa = 3.95WIRR218 pKa = 11.84LFGQGYY224 pKa = 5.09WHH226 pKa = 7.09ISAYY230 pKa = 10.58ALTRR234 pKa = 11.84LLAAPEE240 pKa = 4.13AAKK243 pKa = 10.53SASPPIRR250 pKa = 11.84DD251 pKa = 4.06TILCATT257 pKa = 4.63
MM1 pKa = 7.06TCAKK5 pKa = 9.92VRR7 pKa = 11.84LKK9 pKa = 11.08APLRR13 pKa = 11.84RR14 pKa = 11.84GLTRR18 pKa = 11.84SHH20 pKa = 5.76AHH22 pKa = 6.57KK23 pKa = 10.57SHH25 pKa = 6.07NQDD28 pKa = 2.39HH29 pKa = 5.57VVVVQFWYY37 pKa = 10.17AYY39 pKa = 10.11LPEE42 pKa = 5.24GSSKK46 pKa = 10.46MSWQKK51 pKa = 10.31ISRR54 pKa = 11.84PTVPNQRR61 pKa = 11.84GRR63 pKa = 11.84EE64 pKa = 3.72RR65 pKa = 11.84RR66 pKa = 11.84DD67 pKa = 3.43CIKK70 pKa = 10.38IGSLAAFQNSRR81 pKa = 11.84TISYY85 pKa = 8.53GVPLDD90 pKa = 3.44KK91 pKa = 10.7RR92 pKa = 11.84KK93 pKa = 9.63QRR95 pKa = 11.84AVSSLCLGAAPVDD108 pKa = 3.39QLNCHH113 pKa = 5.93RR114 pKa = 11.84HH115 pKa = 4.44LHH117 pKa = 5.61SRR119 pKa = 11.84PLPRR123 pKa = 11.84RR124 pKa = 11.84NRR126 pKa = 11.84MEE128 pKa = 4.59RR129 pKa = 11.84GPDD132 pKa = 3.19QKK134 pKa = 10.7STLNRR139 pKa = 11.84IRR141 pKa = 11.84LNLKK145 pKa = 8.7SVRR148 pKa = 11.84TCATLASGRR157 pKa = 11.84PIPNLLSPLITLFCADD173 pKa = 4.47HH174 pKa = 6.74SDD176 pKa = 3.94CEE178 pKa = 4.3KK179 pKa = 11.01ASTRR183 pKa = 11.84TEE185 pKa = 3.87RR186 pKa = 11.84LLPDD190 pKa = 3.68PTAPHH195 pKa = 7.49PSRR198 pKa = 11.84FHH200 pKa = 6.7PKK202 pKa = 9.76RR203 pKa = 11.84EE204 pKa = 4.0QVSDD208 pKa = 4.0DD209 pKa = 3.54SATARR214 pKa = 11.84DD215 pKa = 3.95WIRR218 pKa = 11.84LFGQGYY224 pKa = 5.09WHH226 pKa = 7.09ISAYY230 pKa = 10.58ALTRR234 pKa = 11.84LLAAPEE240 pKa = 4.13AAKK243 pKa = 10.53SASPPIRR250 pKa = 11.84DD251 pKa = 4.06TILCATT257 pKa = 4.63
Molecular weight: 28.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1201575 |
40 |
2970 |
313.0 |
34.01 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.066 ± 0.051 | 0.884 ± 0.011 |
6.044 ± 0.047 | 5.722 ± 0.04 |
3.702 ± 0.024 | 8.598 ± 0.051 |
2.11 ± 0.022 | 5.194 ± 0.031 |
3.348 ± 0.033 | 10.131 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.823 ± 0.021 | 2.659 ± 0.025 |
5.016 ± 0.033 | 3.396 ± 0.027 |
6.583 ± 0.049 | 5.227 ± 0.03 |
5.635 ± 0.032 | 7.259 ± 0.035 |
1.354 ± 0.014 | 2.25 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
