
Wenling narna-like virus 1
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.08
Get precalculated fractions of proteins
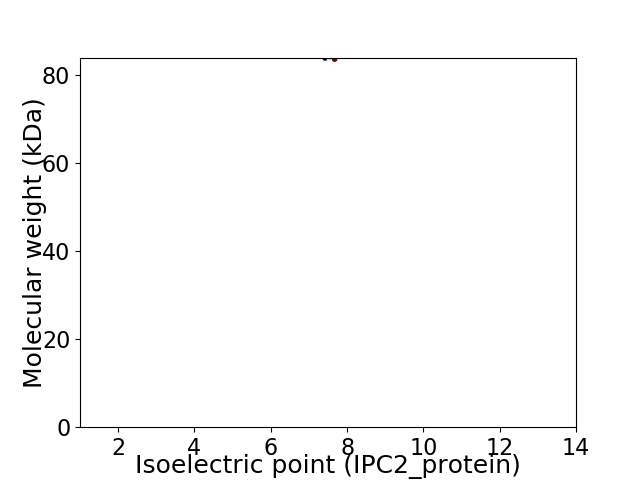
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
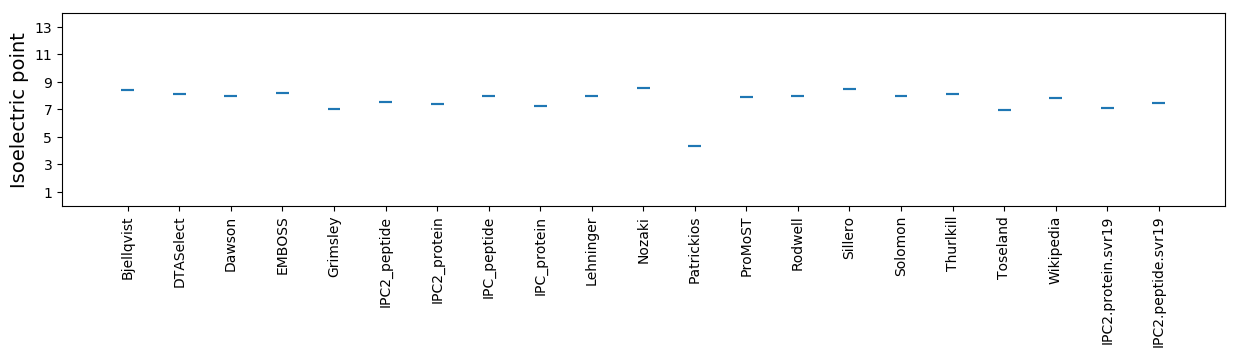
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KIM1|A0A1L3KIM1_9VIRU RNA-dependent RNA polymerase OS=Wenling narna-like virus 1 OX=1923501 PE=4 SV=1
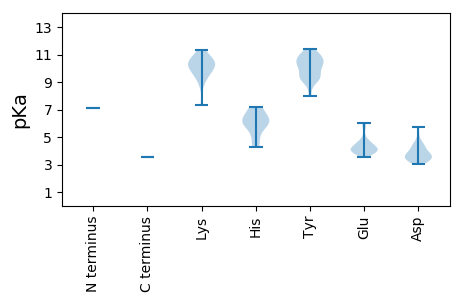
MM1 pKa = 7.1EE2 pKa = 4.43KK3 pKa = 10.39VYY5 pKa = 10.91RR6 pKa = 11.84FLYY9 pKa = 9.1SQYY12 pKa = 10.95GRR14 pKa = 11.84DD15 pKa = 3.53FRR17 pKa = 11.84LSDD20 pKa = 3.65RR21 pKa = 11.84VRR23 pKa = 11.84SIWMYY28 pKa = 10.94FGMLGLLEE36 pKa = 5.85KK37 pKa = 10.22YY38 pKa = 10.28LKK40 pKa = 10.12WKK42 pKa = 8.75TADD45 pKa = 3.34LLAAVWRR52 pKa = 11.84SEE54 pKa = 4.0RR55 pKa = 11.84PPPPSFLTLGLSDD68 pKa = 5.1VPLCLLGTDD77 pKa = 3.82FRR79 pKa = 11.84FWKK82 pKa = 9.73QRR84 pKa = 11.84LRR86 pKa = 11.84SRR88 pKa = 11.84SPTSKK93 pKa = 10.22HH94 pKa = 4.28VQLAFALNQGKK105 pKa = 9.12AAGLPVSDD113 pKa = 4.67DD114 pKa = 3.67FVRR117 pKa = 11.84AAVSSCVEE125 pKa = 4.12RR126 pKa = 11.84LCSDD130 pKa = 3.23EE131 pKa = 6.0DD132 pKa = 3.47IVEE135 pKa = 4.17EE136 pKa = 4.24VTVDD140 pKa = 3.32GTTVTVADD148 pKa = 4.22LRR150 pKa = 11.84RR151 pKa = 11.84EE152 pKa = 3.59ISRR155 pKa = 11.84TVEE158 pKa = 3.81EE159 pKa = 4.74IYY161 pKa = 11.01GHH163 pKa = 5.92GRR165 pKa = 11.84QEE167 pKa = 4.0RR168 pKa = 11.84GTPGSRR174 pKa = 11.84RR175 pKa = 11.84AASVKK180 pKa = 10.51SSVEE184 pKa = 3.9SSRR187 pKa = 11.84SRR189 pKa = 11.84GGPHH193 pKa = 5.99QFLSTQLSTVRR204 pKa = 11.84GSEE207 pKa = 3.97FLVGVAARR215 pKa = 11.84GHH217 pKa = 6.12RR218 pKa = 11.84IVWIHH223 pKa = 6.63APGHH227 pKa = 5.9PEE229 pKa = 3.82DD230 pKa = 4.48WEE232 pKa = 4.24DD233 pKa = 3.86TLWDD237 pKa = 3.65SRR239 pKa = 11.84SRR241 pKa = 11.84AITCQGDD248 pKa = 4.33RR249 pKa = 11.84IPAYY253 pKa = 9.31PVGLTEE259 pKa = 4.05PFKK262 pKa = 11.33VRR264 pKa = 11.84TITRR268 pKa = 11.84GAVDD272 pKa = 4.53PYY274 pKa = 10.56QLARR278 pKa = 11.84RR279 pKa = 11.84WQPSLWKK286 pKa = 9.35PLQRR290 pKa = 11.84HH291 pKa = 4.42EE292 pKa = 4.24CSRR295 pKa = 11.84LVGEE299 pKa = 4.59PVSSGVITSFLGKK312 pKa = 9.86CDD314 pKa = 3.97PEE316 pKa = 4.81DD317 pKa = 3.25GRR319 pKa = 11.84FFVSGDD325 pKa = 3.28YY326 pKa = 10.37EE327 pKa = 4.16AATDD331 pKa = 3.9YY332 pKa = 11.4LNPEE336 pKa = 4.13LSEE339 pKa = 4.23HH340 pKa = 6.27CLEE343 pKa = 4.54EE344 pKa = 4.09VCHH347 pKa = 6.54RR348 pKa = 11.84LGVPPEE354 pKa = 4.02EE355 pKa = 3.97TLVLLKK361 pKa = 10.47CLPRR365 pKa = 11.84HH366 pKa = 5.43EE367 pKa = 5.91LIDD370 pKa = 3.99PVTKK374 pKa = 9.45EE375 pKa = 3.8TLGVQRR381 pKa = 11.84RR382 pKa = 11.84GQLMGSPISFPVLTILNLAFTRR404 pKa = 11.84YY405 pKa = 9.63SLEE408 pKa = 4.68LGACPEE414 pKa = 4.49NPLSTARR421 pKa = 11.84PLDD424 pKa = 3.49DD425 pKa = 5.76HH426 pKa = 7.15EE427 pKa = 4.8ILVNGDD433 pKa = 3.49DD434 pKa = 3.89VLFRR438 pKa = 11.84AHH440 pKa = 6.27PWEE443 pKa = 3.77YY444 pKa = 10.26RR445 pKa = 11.84YY446 pKa = 9.52WKK448 pKa = 10.62AVVALGGLRR457 pKa = 11.84PSLGKK462 pKa = 9.61NQVSRR467 pKa = 11.84RR468 pKa = 11.84YY469 pKa = 8.01FTINSEE475 pKa = 3.84LWHH478 pKa = 7.18AILQPEE484 pKa = 4.55SEE486 pKa = 4.64LFPYY490 pKa = 9.31TWFKK494 pKa = 10.89SEE496 pKa = 4.24RR497 pKa = 11.84LRR499 pKa = 11.84FPQMGLAFGSVKK511 pKa = 10.48GGCSEE516 pKa = 5.8LEE518 pKa = 4.03EE519 pKa = 4.39QPLFSQGAPAAASSRR534 pKa = 11.84TSCWRR539 pKa = 11.84EE540 pKa = 3.58FLQDD544 pKa = 3.56APIPSLAWDD553 pKa = 4.13MLWDD557 pKa = 4.39LSSSLRR563 pKa = 11.84SQVPKK568 pKa = 10.92GMAYY572 pKa = 10.25CLPEE576 pKa = 3.83WLGGLGLPLPPKK588 pKa = 9.78GHH590 pKa = 7.15PLRR593 pKa = 11.84EE594 pKa = 3.7QRR596 pKa = 11.84MAGSASLALARR607 pKa = 11.84YY608 pKa = 9.17IRR610 pKa = 11.84DD611 pKa = 3.15NWDD614 pKa = 3.01TRR616 pKa = 11.84TVRR619 pKa = 11.84SWWRR623 pKa = 11.84GRR625 pKa = 11.84AYY627 pKa = 10.45GGNSPAYY634 pKa = 10.18LEE636 pKa = 4.38YY637 pKa = 10.81ADD639 pKa = 4.37EE640 pKa = 4.1QLHH643 pKa = 6.25RR644 pKa = 11.84ALHH647 pKa = 4.99SLKK650 pKa = 10.45VLPEE654 pKa = 4.09PVPIDD659 pKa = 3.59QDD661 pKa = 3.4PDD663 pKa = 3.64RR664 pKa = 11.84YY665 pKa = 9.74PLPLSSFFPIGVFSYY680 pKa = 9.3EE681 pKa = 4.12YY682 pKa = 9.18CTKK685 pKa = 10.71ADD687 pKa = 3.69KK688 pKa = 11.23EE689 pKa = 3.9EE690 pKa = 4.47DD691 pKa = 3.2MGKK694 pKa = 9.9RR695 pKa = 11.84LANLSKK701 pKa = 10.16SWTRR705 pKa = 11.84LLYY708 pKa = 10.21RR709 pKa = 11.84AWGSKK714 pKa = 7.3TLPLHH719 pKa = 6.0GSEE722 pKa = 5.28IEE724 pKa = 4.15YY725 pKa = 10.79NPPKK729 pKa = 10.3TFLSTRR735 pKa = 11.84WITCC739 pKa = 3.57
MM1 pKa = 7.1EE2 pKa = 4.43KK3 pKa = 10.39VYY5 pKa = 10.91RR6 pKa = 11.84FLYY9 pKa = 9.1SQYY12 pKa = 10.95GRR14 pKa = 11.84DD15 pKa = 3.53FRR17 pKa = 11.84LSDD20 pKa = 3.65RR21 pKa = 11.84VRR23 pKa = 11.84SIWMYY28 pKa = 10.94FGMLGLLEE36 pKa = 5.85KK37 pKa = 10.22YY38 pKa = 10.28LKK40 pKa = 10.12WKK42 pKa = 8.75TADD45 pKa = 3.34LLAAVWRR52 pKa = 11.84SEE54 pKa = 4.0RR55 pKa = 11.84PPPPSFLTLGLSDD68 pKa = 5.1VPLCLLGTDD77 pKa = 3.82FRR79 pKa = 11.84FWKK82 pKa = 9.73QRR84 pKa = 11.84LRR86 pKa = 11.84SRR88 pKa = 11.84SPTSKK93 pKa = 10.22HH94 pKa = 4.28VQLAFALNQGKK105 pKa = 9.12AAGLPVSDD113 pKa = 4.67DD114 pKa = 3.67FVRR117 pKa = 11.84AAVSSCVEE125 pKa = 4.12RR126 pKa = 11.84LCSDD130 pKa = 3.23EE131 pKa = 6.0DD132 pKa = 3.47IVEE135 pKa = 4.17EE136 pKa = 4.24VTVDD140 pKa = 3.32GTTVTVADD148 pKa = 4.22LRR150 pKa = 11.84RR151 pKa = 11.84EE152 pKa = 3.59ISRR155 pKa = 11.84TVEE158 pKa = 3.81EE159 pKa = 4.74IYY161 pKa = 11.01GHH163 pKa = 5.92GRR165 pKa = 11.84QEE167 pKa = 4.0RR168 pKa = 11.84GTPGSRR174 pKa = 11.84RR175 pKa = 11.84AASVKK180 pKa = 10.51SSVEE184 pKa = 3.9SSRR187 pKa = 11.84SRR189 pKa = 11.84GGPHH193 pKa = 5.99QFLSTQLSTVRR204 pKa = 11.84GSEE207 pKa = 3.97FLVGVAARR215 pKa = 11.84GHH217 pKa = 6.12RR218 pKa = 11.84IVWIHH223 pKa = 6.63APGHH227 pKa = 5.9PEE229 pKa = 3.82DD230 pKa = 4.48WEE232 pKa = 4.24DD233 pKa = 3.86TLWDD237 pKa = 3.65SRR239 pKa = 11.84SRR241 pKa = 11.84AITCQGDD248 pKa = 4.33RR249 pKa = 11.84IPAYY253 pKa = 9.31PVGLTEE259 pKa = 4.05PFKK262 pKa = 11.33VRR264 pKa = 11.84TITRR268 pKa = 11.84GAVDD272 pKa = 4.53PYY274 pKa = 10.56QLARR278 pKa = 11.84RR279 pKa = 11.84WQPSLWKK286 pKa = 9.35PLQRR290 pKa = 11.84HH291 pKa = 4.42EE292 pKa = 4.24CSRR295 pKa = 11.84LVGEE299 pKa = 4.59PVSSGVITSFLGKK312 pKa = 9.86CDD314 pKa = 3.97PEE316 pKa = 4.81DD317 pKa = 3.25GRR319 pKa = 11.84FFVSGDD325 pKa = 3.28YY326 pKa = 10.37EE327 pKa = 4.16AATDD331 pKa = 3.9YY332 pKa = 11.4LNPEE336 pKa = 4.13LSEE339 pKa = 4.23HH340 pKa = 6.27CLEE343 pKa = 4.54EE344 pKa = 4.09VCHH347 pKa = 6.54RR348 pKa = 11.84LGVPPEE354 pKa = 4.02EE355 pKa = 3.97TLVLLKK361 pKa = 10.47CLPRR365 pKa = 11.84HH366 pKa = 5.43EE367 pKa = 5.91LIDD370 pKa = 3.99PVTKK374 pKa = 9.45EE375 pKa = 3.8TLGVQRR381 pKa = 11.84RR382 pKa = 11.84GQLMGSPISFPVLTILNLAFTRR404 pKa = 11.84YY405 pKa = 9.63SLEE408 pKa = 4.68LGACPEE414 pKa = 4.49NPLSTARR421 pKa = 11.84PLDD424 pKa = 3.49DD425 pKa = 5.76HH426 pKa = 7.15EE427 pKa = 4.8ILVNGDD433 pKa = 3.49DD434 pKa = 3.89VLFRR438 pKa = 11.84AHH440 pKa = 6.27PWEE443 pKa = 3.77YY444 pKa = 10.26RR445 pKa = 11.84YY446 pKa = 9.52WKK448 pKa = 10.62AVVALGGLRR457 pKa = 11.84PSLGKK462 pKa = 9.61NQVSRR467 pKa = 11.84RR468 pKa = 11.84YY469 pKa = 8.01FTINSEE475 pKa = 3.84LWHH478 pKa = 7.18AILQPEE484 pKa = 4.55SEE486 pKa = 4.64LFPYY490 pKa = 9.31TWFKK494 pKa = 10.89SEE496 pKa = 4.24RR497 pKa = 11.84LRR499 pKa = 11.84FPQMGLAFGSVKK511 pKa = 10.48GGCSEE516 pKa = 5.8LEE518 pKa = 4.03EE519 pKa = 4.39QPLFSQGAPAAASSRR534 pKa = 11.84TSCWRR539 pKa = 11.84EE540 pKa = 3.58FLQDD544 pKa = 3.56APIPSLAWDD553 pKa = 4.13MLWDD557 pKa = 4.39LSSSLRR563 pKa = 11.84SQVPKK568 pKa = 10.92GMAYY572 pKa = 10.25CLPEE576 pKa = 3.83WLGGLGLPLPPKK588 pKa = 9.78GHH590 pKa = 7.15PLRR593 pKa = 11.84EE594 pKa = 3.7QRR596 pKa = 11.84MAGSASLALARR607 pKa = 11.84YY608 pKa = 9.17IRR610 pKa = 11.84DD611 pKa = 3.15NWDD614 pKa = 3.01TRR616 pKa = 11.84TVRR619 pKa = 11.84SWWRR623 pKa = 11.84GRR625 pKa = 11.84AYY627 pKa = 10.45GGNSPAYY634 pKa = 10.18LEE636 pKa = 4.38YY637 pKa = 10.81ADD639 pKa = 4.37EE640 pKa = 4.1QLHH643 pKa = 6.25RR644 pKa = 11.84ALHH647 pKa = 4.99SLKK650 pKa = 10.45VLPEE654 pKa = 4.09PVPIDD659 pKa = 3.59QDD661 pKa = 3.4PDD663 pKa = 3.64RR664 pKa = 11.84YY665 pKa = 9.74PLPLSSFFPIGVFSYY680 pKa = 9.3EE681 pKa = 4.12YY682 pKa = 9.18CTKK685 pKa = 10.71ADD687 pKa = 3.69KK688 pKa = 11.23EE689 pKa = 3.9EE690 pKa = 4.47DD691 pKa = 3.2MGKK694 pKa = 9.9RR695 pKa = 11.84LANLSKK701 pKa = 10.16SWTRR705 pKa = 11.84LLYY708 pKa = 10.21RR709 pKa = 11.84AWGSKK714 pKa = 7.3TLPLHH719 pKa = 6.0GSEE722 pKa = 5.28IEE724 pKa = 4.15YY725 pKa = 10.79NPPKK729 pKa = 10.3TFLSTRR735 pKa = 11.84WITCC739 pKa = 3.57
Molecular weight: 83.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KIM1|A0A1L3KIM1_9VIRU RNA-dependent RNA polymerase OS=Wenling narna-like virus 1 OX=1923501 PE=4 SV=1
MM1 pKa = 7.1EE2 pKa = 4.43KK3 pKa = 10.39VYY5 pKa = 10.91RR6 pKa = 11.84FLYY9 pKa = 9.1SQYY12 pKa = 10.95GRR14 pKa = 11.84DD15 pKa = 3.53FRR17 pKa = 11.84LSDD20 pKa = 3.65RR21 pKa = 11.84VRR23 pKa = 11.84SIWMYY28 pKa = 10.94FGMLGLLEE36 pKa = 5.85KK37 pKa = 10.22YY38 pKa = 10.28LKK40 pKa = 10.12WKK42 pKa = 8.75TADD45 pKa = 3.34LLAAVWRR52 pKa = 11.84SEE54 pKa = 4.0RR55 pKa = 11.84PPPPSFLTLGLSDD68 pKa = 5.1VPLCLLGTDD77 pKa = 3.82FRR79 pKa = 11.84FWKK82 pKa = 9.73QRR84 pKa = 11.84LRR86 pKa = 11.84SRR88 pKa = 11.84SPTSKK93 pKa = 10.22HH94 pKa = 4.28VQLAFALNQGKK105 pKa = 9.12AAGLPVSDD113 pKa = 4.67DD114 pKa = 3.67FVRR117 pKa = 11.84AAVSSCVEE125 pKa = 4.12RR126 pKa = 11.84LCSDD130 pKa = 3.23EE131 pKa = 6.0DD132 pKa = 3.47IVEE135 pKa = 4.17EE136 pKa = 4.24VTVDD140 pKa = 3.32GTTVTVADD148 pKa = 4.22LRR150 pKa = 11.84RR151 pKa = 11.84EE152 pKa = 3.59ISRR155 pKa = 11.84TVEE158 pKa = 3.81EE159 pKa = 4.74IYY161 pKa = 11.01GHH163 pKa = 5.92GRR165 pKa = 11.84QEE167 pKa = 4.0RR168 pKa = 11.84GTPGSRR174 pKa = 11.84RR175 pKa = 11.84AASVKK180 pKa = 10.51SSVEE184 pKa = 3.9SSRR187 pKa = 11.84SRR189 pKa = 11.84GGPHH193 pKa = 5.99QFLSTQLSTVRR204 pKa = 11.84GSEE207 pKa = 3.97FLVGVAARR215 pKa = 11.84GHH217 pKa = 6.12RR218 pKa = 11.84IVWIHH223 pKa = 6.63APGHH227 pKa = 5.9PEE229 pKa = 3.82DD230 pKa = 4.48WEE232 pKa = 4.24DD233 pKa = 3.86TLWDD237 pKa = 3.65SRR239 pKa = 11.84SRR241 pKa = 11.84AITCQGDD248 pKa = 4.33RR249 pKa = 11.84IPAYY253 pKa = 9.31PVGLTEE259 pKa = 4.05PFKK262 pKa = 11.33VRR264 pKa = 11.84TITRR268 pKa = 11.84GAVDD272 pKa = 4.53PYY274 pKa = 10.56QLARR278 pKa = 11.84RR279 pKa = 11.84WQPSLWKK286 pKa = 9.35PLQRR290 pKa = 11.84HH291 pKa = 4.42EE292 pKa = 4.24CSRR295 pKa = 11.84LVGEE299 pKa = 4.59PVSSGVITSFLGKK312 pKa = 9.86CDD314 pKa = 3.97PEE316 pKa = 4.81DD317 pKa = 3.25GRR319 pKa = 11.84FFVSGDD325 pKa = 3.28YY326 pKa = 10.37EE327 pKa = 4.16AATDD331 pKa = 3.9YY332 pKa = 11.4LNPEE336 pKa = 4.13LSEE339 pKa = 4.23HH340 pKa = 6.27CLEE343 pKa = 4.54EE344 pKa = 4.09VCHH347 pKa = 6.54RR348 pKa = 11.84LGVPPEE354 pKa = 4.02EE355 pKa = 3.97TLVLLKK361 pKa = 10.47CLPRR365 pKa = 11.84HH366 pKa = 5.43EE367 pKa = 5.91LIDD370 pKa = 3.99PVTKK374 pKa = 9.45EE375 pKa = 3.8TLGVQRR381 pKa = 11.84RR382 pKa = 11.84GQLMGSPISFPVLTILNLAFTRR404 pKa = 11.84YY405 pKa = 9.63SLEE408 pKa = 4.68LGACPEE414 pKa = 4.49NPLSTARR421 pKa = 11.84PLDD424 pKa = 3.49DD425 pKa = 5.76HH426 pKa = 7.15EE427 pKa = 4.8ILVNGDD433 pKa = 3.49DD434 pKa = 3.89VLFRR438 pKa = 11.84AHH440 pKa = 6.27PWEE443 pKa = 3.77YY444 pKa = 10.26RR445 pKa = 11.84YY446 pKa = 9.52WKK448 pKa = 10.62AVVALGGLRR457 pKa = 11.84PSLGKK462 pKa = 9.61NQVSRR467 pKa = 11.84RR468 pKa = 11.84YY469 pKa = 8.01FTINSEE475 pKa = 3.84LWHH478 pKa = 7.18AILQPEE484 pKa = 4.55SEE486 pKa = 4.64LFPYY490 pKa = 9.31TWFKK494 pKa = 10.89SEE496 pKa = 4.24RR497 pKa = 11.84LRR499 pKa = 11.84FPQMGLAFGSVKK511 pKa = 10.48GGCSEE516 pKa = 5.8LEE518 pKa = 4.03EE519 pKa = 4.39QPLFSQGAPAAASSRR534 pKa = 11.84TSCWRR539 pKa = 11.84EE540 pKa = 3.58FLQDD544 pKa = 3.56APIPSLAWDD553 pKa = 4.13MLWDD557 pKa = 4.39LSSSLRR563 pKa = 11.84SQVPKK568 pKa = 10.92GMAYY572 pKa = 10.25CLPEE576 pKa = 3.83WLGGLGLPLPPKK588 pKa = 9.78GHH590 pKa = 7.15PLRR593 pKa = 11.84EE594 pKa = 3.7QRR596 pKa = 11.84MAGSASLALARR607 pKa = 11.84YY608 pKa = 9.17IRR610 pKa = 11.84DD611 pKa = 3.15NWDD614 pKa = 3.01TRR616 pKa = 11.84TVRR619 pKa = 11.84SWWRR623 pKa = 11.84GRR625 pKa = 11.84AYY627 pKa = 10.45GGNSPAYY634 pKa = 10.18LEE636 pKa = 4.38YY637 pKa = 10.81ADD639 pKa = 4.37EE640 pKa = 4.1QLHH643 pKa = 6.25RR644 pKa = 11.84ALHH647 pKa = 4.99SLKK650 pKa = 10.45VLPEE654 pKa = 4.09PVPIDD659 pKa = 3.59QDD661 pKa = 3.4PDD663 pKa = 3.64RR664 pKa = 11.84YY665 pKa = 9.74PLPLSSFFPIGVFSYY680 pKa = 9.3EE681 pKa = 4.12YY682 pKa = 9.18CTKK685 pKa = 10.71ADD687 pKa = 3.69KK688 pKa = 11.23EE689 pKa = 3.9EE690 pKa = 4.47DD691 pKa = 3.2MGKK694 pKa = 9.9RR695 pKa = 11.84LANLSKK701 pKa = 10.16SWTRR705 pKa = 11.84LLYY708 pKa = 10.21RR709 pKa = 11.84AWGSKK714 pKa = 7.3TLPLHH719 pKa = 6.0GSEE722 pKa = 5.28IEE724 pKa = 4.15YY725 pKa = 10.79NPPKK729 pKa = 10.3TFLSTRR735 pKa = 11.84WITCC739 pKa = 3.57
MM1 pKa = 7.1EE2 pKa = 4.43KK3 pKa = 10.39VYY5 pKa = 10.91RR6 pKa = 11.84FLYY9 pKa = 9.1SQYY12 pKa = 10.95GRR14 pKa = 11.84DD15 pKa = 3.53FRR17 pKa = 11.84LSDD20 pKa = 3.65RR21 pKa = 11.84VRR23 pKa = 11.84SIWMYY28 pKa = 10.94FGMLGLLEE36 pKa = 5.85KK37 pKa = 10.22YY38 pKa = 10.28LKK40 pKa = 10.12WKK42 pKa = 8.75TADD45 pKa = 3.34LLAAVWRR52 pKa = 11.84SEE54 pKa = 4.0RR55 pKa = 11.84PPPPSFLTLGLSDD68 pKa = 5.1VPLCLLGTDD77 pKa = 3.82FRR79 pKa = 11.84FWKK82 pKa = 9.73QRR84 pKa = 11.84LRR86 pKa = 11.84SRR88 pKa = 11.84SPTSKK93 pKa = 10.22HH94 pKa = 4.28VQLAFALNQGKK105 pKa = 9.12AAGLPVSDD113 pKa = 4.67DD114 pKa = 3.67FVRR117 pKa = 11.84AAVSSCVEE125 pKa = 4.12RR126 pKa = 11.84LCSDD130 pKa = 3.23EE131 pKa = 6.0DD132 pKa = 3.47IVEE135 pKa = 4.17EE136 pKa = 4.24VTVDD140 pKa = 3.32GTTVTVADD148 pKa = 4.22LRR150 pKa = 11.84RR151 pKa = 11.84EE152 pKa = 3.59ISRR155 pKa = 11.84TVEE158 pKa = 3.81EE159 pKa = 4.74IYY161 pKa = 11.01GHH163 pKa = 5.92GRR165 pKa = 11.84QEE167 pKa = 4.0RR168 pKa = 11.84GTPGSRR174 pKa = 11.84RR175 pKa = 11.84AASVKK180 pKa = 10.51SSVEE184 pKa = 3.9SSRR187 pKa = 11.84SRR189 pKa = 11.84GGPHH193 pKa = 5.99QFLSTQLSTVRR204 pKa = 11.84GSEE207 pKa = 3.97FLVGVAARR215 pKa = 11.84GHH217 pKa = 6.12RR218 pKa = 11.84IVWIHH223 pKa = 6.63APGHH227 pKa = 5.9PEE229 pKa = 3.82DD230 pKa = 4.48WEE232 pKa = 4.24DD233 pKa = 3.86TLWDD237 pKa = 3.65SRR239 pKa = 11.84SRR241 pKa = 11.84AITCQGDD248 pKa = 4.33RR249 pKa = 11.84IPAYY253 pKa = 9.31PVGLTEE259 pKa = 4.05PFKK262 pKa = 11.33VRR264 pKa = 11.84TITRR268 pKa = 11.84GAVDD272 pKa = 4.53PYY274 pKa = 10.56QLARR278 pKa = 11.84RR279 pKa = 11.84WQPSLWKK286 pKa = 9.35PLQRR290 pKa = 11.84HH291 pKa = 4.42EE292 pKa = 4.24CSRR295 pKa = 11.84LVGEE299 pKa = 4.59PVSSGVITSFLGKK312 pKa = 9.86CDD314 pKa = 3.97PEE316 pKa = 4.81DD317 pKa = 3.25GRR319 pKa = 11.84FFVSGDD325 pKa = 3.28YY326 pKa = 10.37EE327 pKa = 4.16AATDD331 pKa = 3.9YY332 pKa = 11.4LNPEE336 pKa = 4.13LSEE339 pKa = 4.23HH340 pKa = 6.27CLEE343 pKa = 4.54EE344 pKa = 4.09VCHH347 pKa = 6.54RR348 pKa = 11.84LGVPPEE354 pKa = 4.02EE355 pKa = 3.97TLVLLKK361 pKa = 10.47CLPRR365 pKa = 11.84HH366 pKa = 5.43EE367 pKa = 5.91LIDD370 pKa = 3.99PVTKK374 pKa = 9.45EE375 pKa = 3.8TLGVQRR381 pKa = 11.84RR382 pKa = 11.84GQLMGSPISFPVLTILNLAFTRR404 pKa = 11.84YY405 pKa = 9.63SLEE408 pKa = 4.68LGACPEE414 pKa = 4.49NPLSTARR421 pKa = 11.84PLDD424 pKa = 3.49DD425 pKa = 5.76HH426 pKa = 7.15EE427 pKa = 4.8ILVNGDD433 pKa = 3.49DD434 pKa = 3.89VLFRR438 pKa = 11.84AHH440 pKa = 6.27PWEE443 pKa = 3.77YY444 pKa = 10.26RR445 pKa = 11.84YY446 pKa = 9.52WKK448 pKa = 10.62AVVALGGLRR457 pKa = 11.84PSLGKK462 pKa = 9.61NQVSRR467 pKa = 11.84RR468 pKa = 11.84YY469 pKa = 8.01FTINSEE475 pKa = 3.84LWHH478 pKa = 7.18AILQPEE484 pKa = 4.55SEE486 pKa = 4.64LFPYY490 pKa = 9.31TWFKK494 pKa = 10.89SEE496 pKa = 4.24RR497 pKa = 11.84LRR499 pKa = 11.84FPQMGLAFGSVKK511 pKa = 10.48GGCSEE516 pKa = 5.8LEE518 pKa = 4.03EE519 pKa = 4.39QPLFSQGAPAAASSRR534 pKa = 11.84TSCWRR539 pKa = 11.84EE540 pKa = 3.58FLQDD544 pKa = 3.56APIPSLAWDD553 pKa = 4.13MLWDD557 pKa = 4.39LSSSLRR563 pKa = 11.84SQVPKK568 pKa = 10.92GMAYY572 pKa = 10.25CLPEE576 pKa = 3.83WLGGLGLPLPPKK588 pKa = 9.78GHH590 pKa = 7.15PLRR593 pKa = 11.84EE594 pKa = 3.7QRR596 pKa = 11.84MAGSASLALARR607 pKa = 11.84YY608 pKa = 9.17IRR610 pKa = 11.84DD611 pKa = 3.15NWDD614 pKa = 3.01TRR616 pKa = 11.84TVRR619 pKa = 11.84SWWRR623 pKa = 11.84GRR625 pKa = 11.84AYY627 pKa = 10.45GGNSPAYY634 pKa = 10.18LEE636 pKa = 4.38YY637 pKa = 10.81ADD639 pKa = 4.37EE640 pKa = 4.1QLHH643 pKa = 6.25RR644 pKa = 11.84ALHH647 pKa = 4.99SLKK650 pKa = 10.45VLPEE654 pKa = 4.09PVPIDD659 pKa = 3.59QDD661 pKa = 3.4PDD663 pKa = 3.64RR664 pKa = 11.84YY665 pKa = 9.74PLPLSSFFPIGVFSYY680 pKa = 9.3EE681 pKa = 4.12YY682 pKa = 9.18CTKK685 pKa = 10.71ADD687 pKa = 3.69KK688 pKa = 11.23EE689 pKa = 3.9EE690 pKa = 4.47DD691 pKa = 3.2MGKK694 pKa = 9.9RR695 pKa = 11.84LANLSKK701 pKa = 10.16SWTRR705 pKa = 11.84LLYY708 pKa = 10.21RR709 pKa = 11.84AWGSKK714 pKa = 7.3TLPLHH719 pKa = 6.0GSEE722 pKa = 5.28IEE724 pKa = 4.15YY725 pKa = 10.79NPPKK729 pKa = 10.3TFLSTRR735 pKa = 11.84WITCC739 pKa = 3.57
Molecular weight: 83.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
739 |
739 |
739 |
739.0 |
83.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.36 ± 0.0 | 2.03 ± 0.0 |
4.871 ± 0.0 | 6.766 ± 0.0 |
3.789 ± 0.0 | 7.172 ± 0.0 |
2.3 ± 0.0 | 2.977 ± 0.0 |
3.518 ± 0.0 | 11.773 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.218 ± 0.0 | 1.488 ± 0.0 |
7.442 ± 0.0 | 3.112 ± 0.0 |
8.796 ± 0.0 | 8.931 ± 0.0 |
4.871 ± 0.0 | 6.089 ± 0.0 |
3.112 ± 0.0 | 3.383 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
