
Maize yellow mosaic virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus
Average proteome isoelectric point is 8.23
Get precalculated fractions of proteins
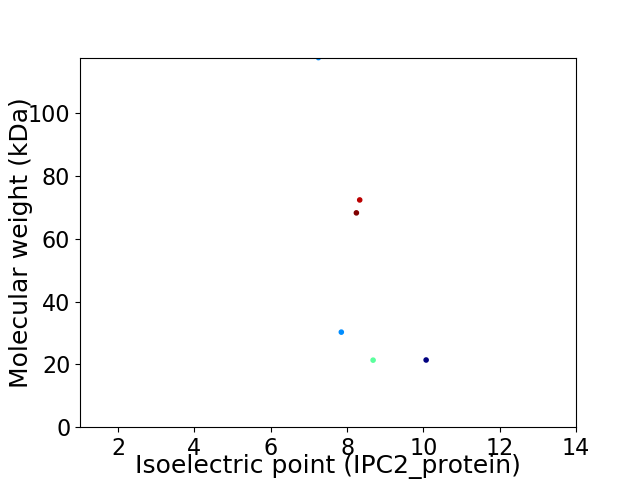
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

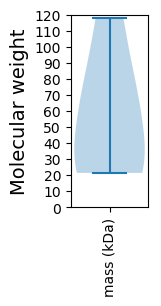
Protein with the lowest isoelectric point:
>tr|A0A191T7R1|A0A191T7R1_9LUTE Coat protein OS=Maize yellow mosaic virus OX=1856642 PE=4 SV=1
MM1 pKa = 7.22QKK3 pKa = 10.27LSSVAFFLCCLLCYY17 pKa = 9.06ATYY20 pKa = 7.68PTRR23 pKa = 11.84GDD25 pKa = 3.53FLEE28 pKa = 4.6MYY30 pKa = 9.86GLPLRR35 pKa = 11.84GTQFGLEE42 pKa = 4.18EE43 pKa = 4.16TSLLPRR49 pKa = 11.84NASLPFKK56 pKa = 10.59FLLAPDD62 pKa = 3.62SCIEE66 pKa = 3.73ISYY69 pKa = 10.17SALSRR74 pKa = 11.84EE75 pKa = 4.26IARR78 pKa = 11.84LALQDD83 pKa = 3.56LHH85 pKa = 6.63TISLAGSRR93 pKa = 11.84ALSSSWKK100 pKa = 10.38GFAEE104 pKa = 4.14EE105 pKa = 4.53WNTSVVKK112 pKa = 10.4LAHH115 pKa = 5.96ATSGFYY121 pKa = 9.9QQASDD126 pKa = 4.03YY127 pKa = 10.88VVSCVLEE134 pKa = 4.0LLGSCLVAIIYY145 pKa = 7.61TWSFFISSLFWVVTFMLQHH164 pKa = 5.16YY165 pKa = 6.74TPHH168 pKa = 5.91VLSIAVLYY176 pKa = 10.43ICTTLLVRR184 pKa = 11.84LATKK188 pKa = 10.33IFGGWPVMMISWIGTSTLGFSKK210 pKa = 9.68TLLFKK215 pKa = 10.9GSSISYY221 pKa = 7.6EE222 pKa = 3.92IPTPGYY228 pKa = 10.11KK229 pKa = 10.02SIEE232 pKa = 4.21IPQKK236 pKa = 9.98PPKK239 pKa = 10.48DD240 pKa = 3.44CVLLVQHH247 pKa = 6.82NDD249 pKa = 3.16ALNSPGGYY257 pKa = 9.68ASCVRR262 pKa = 11.84LLNGSNALLTAEE274 pKa = 4.67HH275 pKa = 6.17VAAQKK280 pKa = 11.34GEE282 pKa = 4.17LLVASTRR289 pKa = 11.84TGNKK293 pKa = 9.14IKK295 pKa = 10.79LSLFNRR301 pKa = 11.84ILTTTNSDD309 pKa = 2.75VSLFQGPPGWEE320 pKa = 3.79SLLGCKK326 pKa = 9.73AADD329 pKa = 3.37ITPVDD334 pKa = 3.66GLTTCEE340 pKa = 3.58ASLFRR345 pKa = 11.84FDD347 pKa = 3.99GHH349 pKa = 5.47WMRR352 pKa = 11.84SNASLVGTEE361 pKa = 3.87EE362 pKa = 4.7TYY364 pKa = 11.51VSVLSNTEE372 pKa = 3.66EE373 pKa = 5.09GYY375 pKa = 10.92SGTPYY380 pKa = 10.68FNGKK384 pKa = 9.77SILGVHH390 pKa = 5.99VGGNATQTNNLMAPIPSIPGLTKK413 pKa = 9.72HH414 pKa = 6.43RR415 pKa = 11.84YY416 pKa = 8.53VFEE419 pKa = 4.19SPQLKK424 pKa = 10.0GRR426 pKa = 11.84LFTEE430 pKa = 4.21QEE432 pKa = 4.17VEE434 pKa = 4.07QLEE437 pKa = 4.18VDD439 pKa = 2.85IDD441 pKa = 3.86EE442 pKa = 4.97AFKK445 pKa = 10.67KK446 pKa = 10.64AYY448 pKa = 10.25DD449 pKa = 3.72LVHH452 pKa = 6.8FKK454 pKa = 11.15SKK456 pKa = 9.38TGRR459 pKa = 11.84NWADD463 pKa = 3.29YY464 pKa = 10.39EE465 pKa = 4.56DD466 pKa = 5.73DD467 pKa = 3.87ISYY470 pKa = 8.15EE471 pKa = 3.93APKK474 pKa = 10.42FQGKK478 pKa = 9.11RR479 pKa = 11.84RR480 pKa = 11.84TRR482 pKa = 11.84FRR484 pKa = 11.84PRR486 pKa = 11.84KK487 pKa = 9.06QNGLAHH493 pKa = 7.32PYY495 pKa = 10.16SLARR499 pKa = 11.84RR500 pKa = 11.84YY501 pKa = 8.86FKK503 pKa = 10.54QRR505 pKa = 11.84YY506 pKa = 6.24SQQGGGSVGQQDD518 pKa = 3.19RR519 pKa = 11.84RR520 pKa = 11.84LSHH523 pKa = 6.74RR524 pKa = 11.84ADD526 pKa = 3.47GCSTNCGISHH536 pKa = 6.56EE537 pKa = 4.61TPKK540 pKa = 10.26EE541 pKa = 3.89QPEE544 pKa = 4.09AAIKK548 pKa = 9.63AQNFASYY555 pKa = 10.39FSSLYY560 pKa = 9.86KK561 pKa = 10.17WEE563 pKa = 4.26VCDD566 pKa = 4.51TRR568 pKa = 11.84SEE570 pKa = 4.45TPGFEE575 pKa = 4.49TCGSLPRR582 pKa = 11.84YY583 pKa = 9.85YY584 pKa = 9.53FTKK587 pKa = 10.4QKK589 pKa = 10.9EE590 pKa = 3.93EE591 pKa = 4.42SEE593 pKa = 4.13WGRR596 pKa = 11.84LLAEE600 pKa = 4.72GNPALAEE607 pKa = 4.16KK608 pKa = 10.7VSGFGWPQFGPAAEE622 pKa = 4.29LKK624 pKa = 10.42SLRR627 pKa = 11.84LQAQRR632 pKa = 11.84WLSRR636 pKa = 11.84AEE638 pKa = 3.99SAKK641 pKa = 10.38IPSAEE646 pKa = 3.78DD647 pKa = 3.1RR648 pKa = 11.84EE649 pKa = 4.24RR650 pKa = 11.84VIRR653 pKa = 11.84KK654 pKa = 6.66TVEE657 pKa = 3.73AYY659 pKa = 7.7KK660 pKa = 9.8TCQTQCPKK668 pKa = 10.3TSQSNLLVWEE678 pKa = 4.38NFLEE682 pKa = 4.3DD683 pKa = 4.04FKK685 pKa = 11.47QAVFSLEE692 pKa = 3.57PDD694 pKa = 3.26AGVGVPFVGYY704 pKa = 10.19DD705 pKa = 3.04KK706 pKa = 9.97RR707 pKa = 11.84THH709 pKa = 6.47RR710 pKa = 11.84GWIEE714 pKa = 4.45DD715 pKa = 3.71PTLLPVLARR724 pKa = 11.84MTFDD728 pKa = 4.25RR729 pKa = 11.84LHH731 pKa = 6.74KK732 pKa = 9.41MSTVKK737 pKa = 10.56FEE739 pKa = 4.37HH740 pKa = 6.11LTAEE744 pKa = 4.34EE745 pKa = 4.14LVQAGLCDD753 pKa = 5.02PIRR756 pKa = 11.84LFVKK760 pKa = 10.72GEE762 pKa = 3.72PHH764 pKa = 6.07KK765 pKa = 10.35QAKK768 pKa = 9.74LDD770 pKa = 3.49EE771 pKa = 4.35GRR773 pKa = 11.84YY774 pKa = 8.74RR775 pKa = 11.84LIMSVSLLDD784 pKa = 3.51QLVARR789 pKa = 11.84VLFQNQNKK797 pKa = 9.66RR798 pKa = 11.84EE799 pKa = 3.93IALWRR804 pKa = 11.84AIPSKK809 pKa = 10.55PGFGLSTDD817 pKa = 3.55DD818 pKa = 3.46QSRR821 pKa = 11.84EE822 pKa = 3.91FVQNLARR829 pKa = 11.84QCGVSSDD836 pKa = 3.95RR837 pKa = 11.84LLQEE841 pKa = 4.21WQLHH845 pKa = 4.78TVPTDD850 pKa = 3.2CSGFDD855 pKa = 3.21WSVAEE860 pKa = 5.47WMLQDD865 pKa = 4.67DD866 pKa = 4.11MEE868 pKa = 4.39VRR870 pKa = 11.84NRR872 pKa = 11.84LTRR875 pKa = 11.84NNTDD879 pKa = 2.87LTRR882 pKa = 11.84RR883 pKa = 11.84LRR885 pKa = 11.84ACWLKK890 pKa = 10.91CISNSVLCLSDD901 pKa = 3.31GTLLAQRR908 pKa = 11.84VAGVQKK914 pKa = 10.39SGSYY918 pKa = 8.25NTSSSNSRR926 pKa = 11.84IRR928 pKa = 11.84VMAAYY933 pKa = 9.93HH934 pKa = 6.74CGATWAMAMGDD945 pKa = 3.95DD946 pKa = 4.15ALEE949 pKa = 4.37SVDD952 pKa = 3.62TCLDD956 pKa = 3.43VYY958 pKa = 11.17KK959 pKa = 10.94DD960 pKa = 3.43LGFKK964 pKa = 10.81VEE966 pKa = 4.03VSKK969 pKa = 10.95QLEE972 pKa = 4.41FCSHH976 pKa = 4.85VFEE979 pKa = 5.34KK980 pKa = 10.75EE981 pKa = 3.64HH982 pKa = 6.75LARR985 pKa = 11.84PVNQNKK991 pKa = 9.32MIYY994 pKa = 9.64KK995 pKa = 10.2LVYY998 pKa = 9.95GYY1000 pKa = 11.16NPANGSSEE1008 pKa = 4.05VLQRR1012 pKa = 11.84YY1013 pKa = 9.24LDD1015 pKa = 3.37ACMSVLHH1022 pKa = 6.23EE1023 pKa = 4.64LRR1025 pKa = 11.84HH1026 pKa = 6.05DD1027 pKa = 4.05PEE1029 pKa = 4.66TVEE1032 pKa = 6.24LLYY1035 pKa = 10.63KK1036 pKa = 10.04WLVSPVQQQKK1046 pKa = 8.01VV1047 pKa = 3.19
MM1 pKa = 7.22QKK3 pKa = 10.27LSSVAFFLCCLLCYY17 pKa = 9.06ATYY20 pKa = 7.68PTRR23 pKa = 11.84GDD25 pKa = 3.53FLEE28 pKa = 4.6MYY30 pKa = 9.86GLPLRR35 pKa = 11.84GTQFGLEE42 pKa = 4.18EE43 pKa = 4.16TSLLPRR49 pKa = 11.84NASLPFKK56 pKa = 10.59FLLAPDD62 pKa = 3.62SCIEE66 pKa = 3.73ISYY69 pKa = 10.17SALSRR74 pKa = 11.84EE75 pKa = 4.26IARR78 pKa = 11.84LALQDD83 pKa = 3.56LHH85 pKa = 6.63TISLAGSRR93 pKa = 11.84ALSSSWKK100 pKa = 10.38GFAEE104 pKa = 4.14EE105 pKa = 4.53WNTSVVKK112 pKa = 10.4LAHH115 pKa = 5.96ATSGFYY121 pKa = 9.9QQASDD126 pKa = 4.03YY127 pKa = 10.88VVSCVLEE134 pKa = 4.0LLGSCLVAIIYY145 pKa = 7.61TWSFFISSLFWVVTFMLQHH164 pKa = 5.16YY165 pKa = 6.74TPHH168 pKa = 5.91VLSIAVLYY176 pKa = 10.43ICTTLLVRR184 pKa = 11.84LATKK188 pKa = 10.33IFGGWPVMMISWIGTSTLGFSKK210 pKa = 9.68TLLFKK215 pKa = 10.9GSSISYY221 pKa = 7.6EE222 pKa = 3.92IPTPGYY228 pKa = 10.11KK229 pKa = 10.02SIEE232 pKa = 4.21IPQKK236 pKa = 9.98PPKK239 pKa = 10.48DD240 pKa = 3.44CVLLVQHH247 pKa = 6.82NDD249 pKa = 3.16ALNSPGGYY257 pKa = 9.68ASCVRR262 pKa = 11.84LLNGSNALLTAEE274 pKa = 4.67HH275 pKa = 6.17VAAQKK280 pKa = 11.34GEE282 pKa = 4.17LLVASTRR289 pKa = 11.84TGNKK293 pKa = 9.14IKK295 pKa = 10.79LSLFNRR301 pKa = 11.84ILTTTNSDD309 pKa = 2.75VSLFQGPPGWEE320 pKa = 3.79SLLGCKK326 pKa = 9.73AADD329 pKa = 3.37ITPVDD334 pKa = 3.66GLTTCEE340 pKa = 3.58ASLFRR345 pKa = 11.84FDD347 pKa = 3.99GHH349 pKa = 5.47WMRR352 pKa = 11.84SNASLVGTEE361 pKa = 3.87EE362 pKa = 4.7TYY364 pKa = 11.51VSVLSNTEE372 pKa = 3.66EE373 pKa = 5.09GYY375 pKa = 10.92SGTPYY380 pKa = 10.68FNGKK384 pKa = 9.77SILGVHH390 pKa = 5.99VGGNATQTNNLMAPIPSIPGLTKK413 pKa = 9.72HH414 pKa = 6.43RR415 pKa = 11.84YY416 pKa = 8.53VFEE419 pKa = 4.19SPQLKK424 pKa = 10.0GRR426 pKa = 11.84LFTEE430 pKa = 4.21QEE432 pKa = 4.17VEE434 pKa = 4.07QLEE437 pKa = 4.18VDD439 pKa = 2.85IDD441 pKa = 3.86EE442 pKa = 4.97AFKK445 pKa = 10.67KK446 pKa = 10.64AYY448 pKa = 10.25DD449 pKa = 3.72LVHH452 pKa = 6.8FKK454 pKa = 11.15SKK456 pKa = 9.38TGRR459 pKa = 11.84NWADD463 pKa = 3.29YY464 pKa = 10.39EE465 pKa = 4.56DD466 pKa = 5.73DD467 pKa = 3.87ISYY470 pKa = 8.15EE471 pKa = 3.93APKK474 pKa = 10.42FQGKK478 pKa = 9.11RR479 pKa = 11.84RR480 pKa = 11.84TRR482 pKa = 11.84FRR484 pKa = 11.84PRR486 pKa = 11.84KK487 pKa = 9.06QNGLAHH493 pKa = 7.32PYY495 pKa = 10.16SLARR499 pKa = 11.84RR500 pKa = 11.84YY501 pKa = 8.86FKK503 pKa = 10.54QRR505 pKa = 11.84YY506 pKa = 6.24SQQGGGSVGQQDD518 pKa = 3.19RR519 pKa = 11.84RR520 pKa = 11.84LSHH523 pKa = 6.74RR524 pKa = 11.84ADD526 pKa = 3.47GCSTNCGISHH536 pKa = 6.56EE537 pKa = 4.61TPKK540 pKa = 10.26EE541 pKa = 3.89QPEE544 pKa = 4.09AAIKK548 pKa = 9.63AQNFASYY555 pKa = 10.39FSSLYY560 pKa = 9.86KK561 pKa = 10.17WEE563 pKa = 4.26VCDD566 pKa = 4.51TRR568 pKa = 11.84SEE570 pKa = 4.45TPGFEE575 pKa = 4.49TCGSLPRR582 pKa = 11.84YY583 pKa = 9.85YY584 pKa = 9.53FTKK587 pKa = 10.4QKK589 pKa = 10.9EE590 pKa = 3.93EE591 pKa = 4.42SEE593 pKa = 4.13WGRR596 pKa = 11.84LLAEE600 pKa = 4.72GNPALAEE607 pKa = 4.16KK608 pKa = 10.7VSGFGWPQFGPAAEE622 pKa = 4.29LKK624 pKa = 10.42SLRR627 pKa = 11.84LQAQRR632 pKa = 11.84WLSRR636 pKa = 11.84AEE638 pKa = 3.99SAKK641 pKa = 10.38IPSAEE646 pKa = 3.78DD647 pKa = 3.1RR648 pKa = 11.84EE649 pKa = 4.24RR650 pKa = 11.84VIRR653 pKa = 11.84KK654 pKa = 6.66TVEE657 pKa = 3.73AYY659 pKa = 7.7KK660 pKa = 9.8TCQTQCPKK668 pKa = 10.3TSQSNLLVWEE678 pKa = 4.38NFLEE682 pKa = 4.3DD683 pKa = 4.04FKK685 pKa = 11.47QAVFSLEE692 pKa = 3.57PDD694 pKa = 3.26AGVGVPFVGYY704 pKa = 10.19DD705 pKa = 3.04KK706 pKa = 9.97RR707 pKa = 11.84THH709 pKa = 6.47RR710 pKa = 11.84GWIEE714 pKa = 4.45DD715 pKa = 3.71PTLLPVLARR724 pKa = 11.84MTFDD728 pKa = 4.25RR729 pKa = 11.84LHH731 pKa = 6.74KK732 pKa = 9.41MSTVKK737 pKa = 10.56FEE739 pKa = 4.37HH740 pKa = 6.11LTAEE744 pKa = 4.34EE745 pKa = 4.14LVQAGLCDD753 pKa = 5.02PIRR756 pKa = 11.84LFVKK760 pKa = 10.72GEE762 pKa = 3.72PHH764 pKa = 6.07KK765 pKa = 10.35QAKK768 pKa = 9.74LDD770 pKa = 3.49EE771 pKa = 4.35GRR773 pKa = 11.84YY774 pKa = 8.74RR775 pKa = 11.84LIMSVSLLDD784 pKa = 3.51QLVARR789 pKa = 11.84VLFQNQNKK797 pKa = 9.66RR798 pKa = 11.84EE799 pKa = 3.93IALWRR804 pKa = 11.84AIPSKK809 pKa = 10.55PGFGLSTDD817 pKa = 3.55DD818 pKa = 3.46QSRR821 pKa = 11.84EE822 pKa = 3.91FVQNLARR829 pKa = 11.84QCGVSSDD836 pKa = 3.95RR837 pKa = 11.84LLQEE841 pKa = 4.21WQLHH845 pKa = 4.78TVPTDD850 pKa = 3.2CSGFDD855 pKa = 3.21WSVAEE860 pKa = 5.47WMLQDD865 pKa = 4.67DD866 pKa = 4.11MEE868 pKa = 4.39VRR870 pKa = 11.84NRR872 pKa = 11.84LTRR875 pKa = 11.84NNTDD879 pKa = 2.87LTRR882 pKa = 11.84RR883 pKa = 11.84LRR885 pKa = 11.84ACWLKK890 pKa = 10.91CISNSVLCLSDD901 pKa = 3.31GTLLAQRR908 pKa = 11.84VAGVQKK914 pKa = 10.39SGSYY918 pKa = 8.25NTSSSNSRR926 pKa = 11.84IRR928 pKa = 11.84VMAAYY933 pKa = 9.93HH934 pKa = 6.74CGATWAMAMGDD945 pKa = 3.95DD946 pKa = 4.15ALEE949 pKa = 4.37SVDD952 pKa = 3.62TCLDD956 pKa = 3.43VYY958 pKa = 11.17KK959 pKa = 10.94DD960 pKa = 3.43LGFKK964 pKa = 10.81VEE966 pKa = 4.03VSKK969 pKa = 10.95QLEE972 pKa = 4.41FCSHH976 pKa = 4.85VFEE979 pKa = 5.34KK980 pKa = 10.75EE981 pKa = 3.64HH982 pKa = 6.75LARR985 pKa = 11.84PVNQNKK991 pKa = 9.32MIYY994 pKa = 9.64KK995 pKa = 10.2LVYY998 pKa = 9.95GYY1000 pKa = 11.16NPANGSSEE1008 pKa = 4.05VLQRR1012 pKa = 11.84YY1013 pKa = 9.24LDD1015 pKa = 3.37ACMSVLHH1022 pKa = 6.23EE1023 pKa = 4.64LRR1025 pKa = 11.84HH1026 pKa = 6.05DD1027 pKa = 4.05PEE1029 pKa = 4.66TVEE1032 pKa = 6.24LLYY1035 pKa = 10.63KK1036 pKa = 10.04WLVSPVQQQKK1046 pKa = 8.01VV1047 pKa = 3.19
Molecular weight: 117.67 kDa
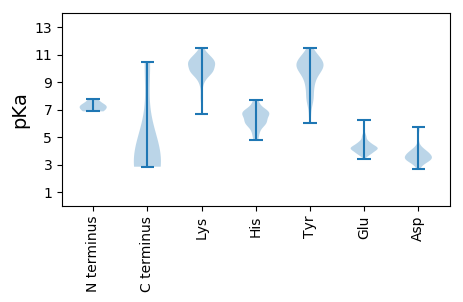
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A191T7S4|A0A191T7S4_9LUTE Movement protein OS=Maize yellow mosaic virus OX=1856642 PE=3 SV=1
MM1 pKa = 6.87NTGGRR6 pKa = 11.84NGRR9 pKa = 11.84RR10 pKa = 11.84ARR12 pKa = 11.84NRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84ARR18 pKa = 11.84NNNRR22 pKa = 11.84AQPVVVVAANPRR34 pKa = 11.84RR35 pKa = 11.84GRR37 pKa = 11.84PRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84PSGNTAGRR51 pKa = 11.84PGVRR55 pKa = 11.84RR56 pKa = 11.84GSRR59 pKa = 11.84EE60 pKa = 3.68TFVFSKK66 pKa = 11.02DD67 pKa = 3.43SLTGNASGKK76 pKa = 7.79VTFGPSLSEE85 pKa = 3.84CAAFSGGILKK95 pKa = 10.27AYY97 pKa = 10.21HH98 pKa = 6.75EE99 pKa = 4.41YY100 pKa = 10.44KK101 pKa = 9.82ISKK104 pKa = 9.83IILEE108 pKa = 5.09FISEE112 pKa = 4.21AASTAEE118 pKa = 3.73GSIAYY123 pKa = 9.5EE124 pKa = 3.8LDD126 pKa = 3.19PHH128 pKa = 6.74NKK130 pKa = 9.68LSSLASTINKK140 pKa = 9.5FSIVKK145 pKa = 9.57GGRR148 pKa = 11.84RR149 pKa = 11.84VFASNQIGGGVWRR162 pKa = 11.84DD163 pKa = 3.37SSEE166 pKa = 4.04DD167 pKa = 3.28QCAILYY173 pKa = 9.54KK174 pKa = 10.95GNGKK178 pKa = 10.21SSVAGSFRR186 pKa = 11.84ITIEE190 pKa = 4.21VNVQNPKK197 pKa = 10.48
MM1 pKa = 6.87NTGGRR6 pKa = 11.84NGRR9 pKa = 11.84RR10 pKa = 11.84ARR12 pKa = 11.84NRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84ARR18 pKa = 11.84NNNRR22 pKa = 11.84AQPVVVVAANPRR34 pKa = 11.84RR35 pKa = 11.84GRR37 pKa = 11.84PRR39 pKa = 11.84RR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84PSGNTAGRR51 pKa = 11.84PGVRR55 pKa = 11.84RR56 pKa = 11.84GSRR59 pKa = 11.84EE60 pKa = 3.68TFVFSKK66 pKa = 11.02DD67 pKa = 3.43SLTGNASGKK76 pKa = 7.79VTFGPSLSEE85 pKa = 3.84CAAFSGGILKK95 pKa = 10.27AYY97 pKa = 10.21HH98 pKa = 6.75EE99 pKa = 4.41YY100 pKa = 10.44KK101 pKa = 9.82ISKK104 pKa = 9.83IILEE108 pKa = 5.09FISEE112 pKa = 4.21AASTAEE118 pKa = 3.73GSIAYY123 pKa = 9.5EE124 pKa = 3.8LDD126 pKa = 3.19PHH128 pKa = 6.74NKK130 pKa = 9.68LSSLASTINKK140 pKa = 9.5FSIVKK145 pKa = 9.57GGRR148 pKa = 11.84RR149 pKa = 11.84VFASNQIGGGVWRR162 pKa = 11.84DD163 pKa = 3.37SSEE166 pKa = 4.04DD167 pKa = 3.28QCAILYY173 pKa = 9.54KK174 pKa = 10.95GNGKK178 pKa = 10.21SSVAGSFRR186 pKa = 11.84ITIEE190 pKa = 4.21VNVQNPKK197 pKa = 10.48
Molecular weight: 21.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2986 |
192 |
1047 |
497.7 |
55.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.368 ± 0.48 | 1.808 ± 0.367 |
4.086 ± 0.364 | 5.626 ± 0.3 |
4.052 ± 0.491 | 7.167 ± 0.455 |
1.842 ± 0.377 | 4.153 ± 0.329 |
4.789 ± 0.471 | 9.277 ± 1.564 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.44 ± 0.152 | 4.253 ± 0.67 |
5.894 ± 0.847 | 4.22 ± 0.356 |
6.899 ± 1.043 | 9.913 ± 0.699 |
6.095 ± 0.363 | 6.196 ± 0.323 |
1.574 ± 0.236 | 3.315 ± 0.241 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
