
Lutzomyia reovirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; unclassified Reoviridae
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
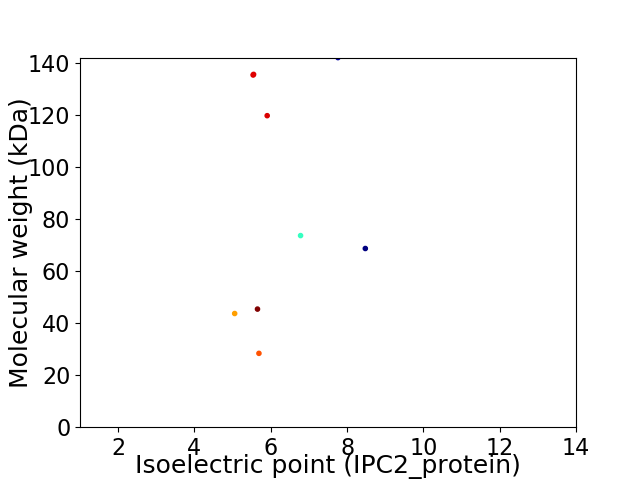
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0H4M970|A0A0H4M970_9REOV RNA-dependent RNA polymerase OS=Lutzomyia reovirus 1 OX=1670669 PE=4 SV=1
MM1 pKa = 7.41MNFDD5 pKa = 4.18STIHH9 pKa = 6.51TIPIFFNLLTVLTTATIDD27 pKa = 3.4RR28 pKa = 11.84DD29 pKa = 3.86GLYY32 pKa = 10.32KK33 pKa = 10.54SYY35 pKa = 10.91ASPVSPSTIPAEE47 pKa = 4.57FIVPLAPPMLQLLNYY62 pKa = 7.26KK63 pKa = 9.25TSPQRR68 pKa = 11.84PLMVLQSFTAALWNSSNYY86 pKa = 9.28HH87 pKa = 5.75RR88 pKa = 11.84LKK90 pKa = 10.3IPRR93 pKa = 11.84SIVDD97 pKa = 3.2ITRR100 pKa = 11.84LASDD104 pKa = 3.5MTQWPWLQLLLDD116 pKa = 4.51CTNNWNYY123 pKa = 9.78TPEE126 pKa = 4.11FKK128 pKa = 10.85ALLVSVCHH136 pKa = 6.66AIEE139 pKa = 3.92IVVIGSVASNIANTPSTYY157 pKa = 10.31EE158 pKa = 4.02FEE160 pKa = 4.61SPSYY164 pKa = 9.01TLPSTYY170 pKa = 10.75DD171 pKa = 3.49PDD173 pKa = 3.93KK174 pKa = 11.04LTITLPRR181 pKa = 11.84MSMICKK187 pKa = 10.04LLQNILIPALEE198 pKa = 3.92RR199 pKa = 11.84VYY201 pKa = 11.18NIIDD205 pKa = 3.64VVDD208 pKa = 4.13DD209 pKa = 3.73VTPQFPTPPTSPILQEE225 pKa = 4.11SLSDD229 pKa = 3.41EE230 pKa = 4.98DD231 pKa = 4.03EE232 pKa = 4.28ATNTDD237 pKa = 2.82VRR239 pKa = 11.84YY240 pKa = 8.81QAYY243 pKa = 8.64YY244 pKa = 9.62AQRR247 pKa = 11.84ALSRR251 pKa = 11.84ICDD254 pKa = 3.89QITDD258 pKa = 5.46AIPCGSHH265 pKa = 7.13LLLSDD270 pKa = 4.4AAPDD274 pKa = 3.49VVIFYY279 pKa = 7.42FTNGNDD285 pKa = 3.32VFIGKK290 pKa = 7.68VEE292 pKa = 4.14RR293 pKa = 11.84SKK295 pKa = 9.9QTTLEE300 pKa = 3.87FEE302 pKa = 5.0YY303 pKa = 10.66IRR305 pKa = 11.84IAHH308 pKa = 6.97IDD310 pKa = 3.6QEE312 pKa = 4.44QAALKK317 pKa = 10.3AIQHH321 pKa = 5.46NVNLFRR327 pKa = 11.84NQLTQPEE334 pKa = 4.74LRR336 pKa = 11.84TLFSDD341 pKa = 4.11EE342 pKa = 4.37SVSMILSHH350 pKa = 5.51THH352 pKa = 6.69FIYY355 pKa = 10.38IPIHH359 pKa = 5.25EE360 pKa = 4.71RR361 pKa = 11.84PLRR364 pKa = 11.84EE365 pKa = 4.22IINGVIASVALIPSQALVTSKK386 pKa = 11.13GSS388 pKa = 3.15
MM1 pKa = 7.41MNFDD5 pKa = 4.18STIHH9 pKa = 6.51TIPIFFNLLTVLTTATIDD27 pKa = 3.4RR28 pKa = 11.84DD29 pKa = 3.86GLYY32 pKa = 10.32KK33 pKa = 10.54SYY35 pKa = 10.91ASPVSPSTIPAEE47 pKa = 4.57FIVPLAPPMLQLLNYY62 pKa = 7.26KK63 pKa = 9.25TSPQRR68 pKa = 11.84PLMVLQSFTAALWNSSNYY86 pKa = 9.28HH87 pKa = 5.75RR88 pKa = 11.84LKK90 pKa = 10.3IPRR93 pKa = 11.84SIVDD97 pKa = 3.2ITRR100 pKa = 11.84LASDD104 pKa = 3.5MTQWPWLQLLLDD116 pKa = 4.51CTNNWNYY123 pKa = 9.78TPEE126 pKa = 4.11FKK128 pKa = 10.85ALLVSVCHH136 pKa = 6.66AIEE139 pKa = 3.92IVVIGSVASNIANTPSTYY157 pKa = 10.31EE158 pKa = 4.02FEE160 pKa = 4.61SPSYY164 pKa = 9.01TLPSTYY170 pKa = 10.75DD171 pKa = 3.49PDD173 pKa = 3.93KK174 pKa = 11.04LTITLPRR181 pKa = 11.84MSMICKK187 pKa = 10.04LLQNILIPALEE198 pKa = 3.92RR199 pKa = 11.84VYY201 pKa = 11.18NIIDD205 pKa = 3.64VVDD208 pKa = 4.13DD209 pKa = 3.73VTPQFPTPPTSPILQEE225 pKa = 4.11SLSDD229 pKa = 3.41EE230 pKa = 4.98DD231 pKa = 4.03EE232 pKa = 4.28ATNTDD237 pKa = 2.82VRR239 pKa = 11.84YY240 pKa = 8.81QAYY243 pKa = 8.64YY244 pKa = 9.62AQRR247 pKa = 11.84ALSRR251 pKa = 11.84ICDD254 pKa = 3.89QITDD258 pKa = 5.46AIPCGSHH265 pKa = 7.13LLLSDD270 pKa = 4.4AAPDD274 pKa = 3.49VVIFYY279 pKa = 7.42FTNGNDD285 pKa = 3.32VFIGKK290 pKa = 7.68VEE292 pKa = 4.14RR293 pKa = 11.84SKK295 pKa = 9.9QTTLEE300 pKa = 3.87FEE302 pKa = 5.0YY303 pKa = 10.66IRR305 pKa = 11.84IAHH308 pKa = 6.97IDD310 pKa = 3.6QEE312 pKa = 4.44QAALKK317 pKa = 10.3AIQHH321 pKa = 5.46NVNLFRR327 pKa = 11.84NQLTQPEE334 pKa = 4.74LRR336 pKa = 11.84TLFSDD341 pKa = 4.11EE342 pKa = 4.37SVSMILSHH350 pKa = 5.51THH352 pKa = 6.69FIYY355 pKa = 10.38IPIHH359 pKa = 5.25EE360 pKa = 4.71RR361 pKa = 11.84PLRR364 pKa = 11.84EE365 pKa = 4.22IINGVIASVALIPSQALVTSKK386 pKa = 11.13GSS388 pKa = 3.15
Molecular weight: 43.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0H4LY56|A0A0H4LY56_9REOV VP1 OS=Lutzomyia reovirus 1 OX=1670669 PE=4 SV=1
MM1 pKa = 7.63EE2 pKa = 5.98LLDD5 pKa = 4.55TEE7 pKa = 5.75DD8 pKa = 3.54STTQHH13 pKa = 5.6IRR15 pKa = 11.84KK16 pKa = 9.0QKK18 pKa = 10.71GLFKK22 pKa = 10.35KK23 pKa = 10.58SKK25 pKa = 9.13ISEE28 pKa = 4.57TITIQQRR35 pKa = 11.84YY36 pKa = 8.4SNIHH40 pKa = 4.88TQHH43 pKa = 5.92KK44 pKa = 8.28TVNLPTNHH52 pKa = 6.42VITLYY57 pKa = 8.68PHH59 pKa = 7.4RR60 pKa = 11.84ITTHH64 pKa = 3.96IHH66 pKa = 4.09QVLRR70 pKa = 11.84KK71 pKa = 9.4IYY73 pKa = 9.97GRR75 pKa = 11.84LDD77 pKa = 3.23AYY79 pKa = 10.02IDD81 pKa = 3.42PRR83 pKa = 11.84QIARR87 pKa = 11.84TLQSGEE93 pKa = 4.22RR94 pKa = 11.84YY95 pKa = 8.81RR96 pKa = 11.84QAVHH100 pKa = 6.44KK101 pKa = 10.05LKK103 pKa = 10.71SKK105 pKa = 10.53RR106 pKa = 11.84IRR108 pKa = 11.84NTSSAKK114 pKa = 6.99QTYY117 pKa = 9.49IFSVDD122 pKa = 3.66DD123 pKa = 4.16KK124 pKa = 11.08LSNDD128 pKa = 3.28ATQEE132 pKa = 3.99EE133 pKa = 5.1LDD135 pKa = 3.8QFQKK139 pKa = 11.05DD140 pKa = 3.97VNHH143 pKa = 6.74LADD146 pKa = 3.8IRR148 pKa = 11.84TQITTGLIFIHH159 pKa = 6.61LGYY162 pKa = 10.35HH163 pKa = 6.79EE164 pKa = 4.7YY165 pKa = 10.62HH166 pKa = 6.9CAIQPYY172 pKa = 8.7MRR174 pKa = 11.84ALMPYY179 pKa = 9.89LPIYY183 pKa = 9.31TCSLVNGFPNYY194 pKa = 10.26ADD196 pKa = 3.91PYY198 pKa = 9.56IANFYY203 pKa = 9.11LAPYY207 pKa = 8.34YY208 pKa = 10.65ASHH211 pKa = 6.54LTDD214 pKa = 4.6AYY216 pKa = 9.49TPKK219 pKa = 9.17PTTRR223 pKa = 11.84FIYY226 pKa = 10.1IDD228 pKa = 3.57TNLTARR234 pKa = 11.84QSIQVKK240 pKa = 9.25RR241 pKa = 11.84HH242 pKa = 5.18IKK244 pKa = 8.34YY245 pKa = 9.85QPSGTLITLSRR256 pKa = 11.84DD257 pKa = 2.97NSLPILFVSTKK268 pKa = 9.56HH269 pKa = 5.8LHH271 pKa = 6.26DD272 pKa = 4.3AKK274 pKa = 10.81EE275 pKa = 4.05YY276 pKa = 10.23IRR278 pKa = 11.84GKK280 pKa = 10.47RR281 pKa = 11.84LIEE284 pKa = 3.75LLYY287 pKa = 10.32DD288 pKa = 3.57YY289 pKa = 10.78SYY291 pKa = 10.0TAPHH295 pKa = 6.56QDD297 pKa = 2.58TTQRR301 pKa = 11.84IDD303 pKa = 3.71IISSAPLLTTITAYY317 pKa = 10.74SPTKK321 pKa = 9.35LQPYY325 pKa = 8.38PPPQCRR331 pKa = 11.84DD332 pKa = 3.02SALTVITKK340 pKa = 9.83FSHH343 pKa = 7.14HH344 pKa = 6.45IQNNHH349 pKa = 5.28QLTAILGVIGTKK361 pKa = 10.68GSGKK365 pKa = 8.07TAGLQRR371 pKa = 11.84AITFMQTTIPWTICHH386 pKa = 6.84IDD388 pKa = 3.15SDD390 pKa = 4.88AYY392 pKa = 10.54GRR394 pKa = 11.84WLAYY398 pKa = 10.35AITSTGTITEE408 pKa = 4.19ITQSFIDD415 pKa = 3.38DD416 pKa = 4.74HH417 pKa = 8.37IYY419 pKa = 11.12NEE421 pKa = 4.85YY422 pKa = 9.83KK423 pKa = 10.23QQDD426 pKa = 3.55NNSPIFHH433 pKa = 6.83EE434 pKa = 5.02LVMQKK439 pKa = 9.74ILNDD443 pKa = 3.92AKK445 pKa = 10.08ITTYY449 pKa = 11.06DD450 pKa = 3.1QFVNLSITQRR460 pKa = 11.84SNKK463 pKa = 9.46IYY465 pKa = 10.45EE466 pKa = 4.52PFTKK470 pKa = 10.23FYY472 pKa = 9.6ITTLSDD478 pKa = 3.3EE479 pKa = 4.83VYY481 pKa = 11.15GLANYY486 pKa = 7.29MTTLLAKK493 pKa = 9.08MDD495 pKa = 3.37HH496 pKa = 6.08RR497 pKa = 11.84VYY499 pKa = 10.38IIEE502 pKa = 4.21CHH504 pKa = 5.96TSVEE508 pKa = 3.91LAKK511 pKa = 10.43AVRR514 pKa = 11.84SDD516 pKa = 3.47TSCILDD522 pKa = 3.55TPFDD526 pKa = 3.65EE527 pKa = 5.17RR528 pKa = 11.84VAITHH533 pKa = 5.88RR534 pKa = 11.84TRR536 pKa = 11.84GDD538 pKa = 3.16KK539 pKa = 10.01TNKK542 pKa = 9.69LSQLVLHH549 pKa = 6.95EE550 pKa = 4.23YY551 pKa = 9.91YY552 pKa = 10.33RR553 pKa = 11.84ARR555 pKa = 11.84IAATIPYY562 pKa = 8.6VRR564 pKa = 11.84LYY566 pKa = 9.91EE567 pKa = 5.98LYY569 pKa = 9.42MCWTEE574 pKa = 4.54LSTSLLHH581 pKa = 6.96KK582 pKa = 10.04GVPQEE587 pKa = 4.15VMTPAPSNN595 pKa = 3.46
MM1 pKa = 7.63EE2 pKa = 5.98LLDD5 pKa = 4.55TEE7 pKa = 5.75DD8 pKa = 3.54STTQHH13 pKa = 5.6IRR15 pKa = 11.84KK16 pKa = 9.0QKK18 pKa = 10.71GLFKK22 pKa = 10.35KK23 pKa = 10.58SKK25 pKa = 9.13ISEE28 pKa = 4.57TITIQQRR35 pKa = 11.84YY36 pKa = 8.4SNIHH40 pKa = 4.88TQHH43 pKa = 5.92KK44 pKa = 8.28TVNLPTNHH52 pKa = 6.42VITLYY57 pKa = 8.68PHH59 pKa = 7.4RR60 pKa = 11.84ITTHH64 pKa = 3.96IHH66 pKa = 4.09QVLRR70 pKa = 11.84KK71 pKa = 9.4IYY73 pKa = 9.97GRR75 pKa = 11.84LDD77 pKa = 3.23AYY79 pKa = 10.02IDD81 pKa = 3.42PRR83 pKa = 11.84QIARR87 pKa = 11.84TLQSGEE93 pKa = 4.22RR94 pKa = 11.84YY95 pKa = 8.81RR96 pKa = 11.84QAVHH100 pKa = 6.44KK101 pKa = 10.05LKK103 pKa = 10.71SKK105 pKa = 10.53RR106 pKa = 11.84IRR108 pKa = 11.84NTSSAKK114 pKa = 6.99QTYY117 pKa = 9.49IFSVDD122 pKa = 3.66DD123 pKa = 4.16KK124 pKa = 11.08LSNDD128 pKa = 3.28ATQEE132 pKa = 3.99EE133 pKa = 5.1LDD135 pKa = 3.8QFQKK139 pKa = 11.05DD140 pKa = 3.97VNHH143 pKa = 6.74LADD146 pKa = 3.8IRR148 pKa = 11.84TQITTGLIFIHH159 pKa = 6.61LGYY162 pKa = 10.35HH163 pKa = 6.79EE164 pKa = 4.7YY165 pKa = 10.62HH166 pKa = 6.9CAIQPYY172 pKa = 8.7MRR174 pKa = 11.84ALMPYY179 pKa = 9.89LPIYY183 pKa = 9.31TCSLVNGFPNYY194 pKa = 10.26ADD196 pKa = 3.91PYY198 pKa = 9.56IANFYY203 pKa = 9.11LAPYY207 pKa = 8.34YY208 pKa = 10.65ASHH211 pKa = 6.54LTDD214 pKa = 4.6AYY216 pKa = 9.49TPKK219 pKa = 9.17PTTRR223 pKa = 11.84FIYY226 pKa = 10.1IDD228 pKa = 3.57TNLTARR234 pKa = 11.84QSIQVKK240 pKa = 9.25RR241 pKa = 11.84HH242 pKa = 5.18IKK244 pKa = 8.34YY245 pKa = 9.85QPSGTLITLSRR256 pKa = 11.84DD257 pKa = 2.97NSLPILFVSTKK268 pKa = 9.56HH269 pKa = 5.8LHH271 pKa = 6.26DD272 pKa = 4.3AKK274 pKa = 10.81EE275 pKa = 4.05YY276 pKa = 10.23IRR278 pKa = 11.84GKK280 pKa = 10.47RR281 pKa = 11.84LIEE284 pKa = 3.75LLYY287 pKa = 10.32DD288 pKa = 3.57YY289 pKa = 10.78SYY291 pKa = 10.0TAPHH295 pKa = 6.56QDD297 pKa = 2.58TTQRR301 pKa = 11.84IDD303 pKa = 3.71IISSAPLLTTITAYY317 pKa = 10.74SPTKK321 pKa = 9.35LQPYY325 pKa = 8.38PPPQCRR331 pKa = 11.84DD332 pKa = 3.02SALTVITKK340 pKa = 9.83FSHH343 pKa = 7.14HH344 pKa = 6.45IQNNHH349 pKa = 5.28QLTAILGVIGTKK361 pKa = 10.68GSGKK365 pKa = 8.07TAGLQRR371 pKa = 11.84AITFMQTTIPWTICHH386 pKa = 6.84IDD388 pKa = 3.15SDD390 pKa = 4.88AYY392 pKa = 10.54GRR394 pKa = 11.84WLAYY398 pKa = 10.35AITSTGTITEE408 pKa = 4.19ITQSFIDD415 pKa = 3.38DD416 pKa = 4.74HH417 pKa = 8.37IYY419 pKa = 11.12NEE421 pKa = 4.85YY422 pKa = 9.83KK423 pKa = 10.23QQDD426 pKa = 3.55NNSPIFHH433 pKa = 6.83EE434 pKa = 5.02LVMQKK439 pKa = 9.74ILNDD443 pKa = 3.92AKK445 pKa = 10.08ITTYY449 pKa = 11.06DD450 pKa = 3.1QFVNLSITQRR460 pKa = 11.84SNKK463 pKa = 9.46IYY465 pKa = 10.45EE466 pKa = 4.52PFTKK470 pKa = 10.23FYY472 pKa = 9.6ITTLSDD478 pKa = 3.3EE479 pKa = 4.83VYY481 pKa = 11.15GLANYY486 pKa = 7.29MTTLLAKK493 pKa = 9.08MDD495 pKa = 3.37HH496 pKa = 6.08RR497 pKa = 11.84VYY499 pKa = 10.38IIEE502 pKa = 4.21CHH504 pKa = 5.96TSVEE508 pKa = 3.91LAKK511 pKa = 10.43AVRR514 pKa = 11.84SDD516 pKa = 3.47TSCILDD522 pKa = 3.55TPFDD526 pKa = 3.65EE527 pKa = 5.17RR528 pKa = 11.84VAITHH533 pKa = 5.88RR534 pKa = 11.84TRR536 pKa = 11.84GDD538 pKa = 3.16KK539 pKa = 10.01TNKK542 pKa = 9.69LSQLVLHH549 pKa = 6.95EE550 pKa = 4.23YY551 pKa = 9.91YY552 pKa = 10.33RR553 pKa = 11.84ARR555 pKa = 11.84IAATIPYY562 pKa = 8.6VRR564 pKa = 11.84LYY566 pKa = 9.91EE567 pKa = 5.98LYY569 pKa = 9.42MCWTEE574 pKa = 4.54LSTSLLHH581 pKa = 6.96KK582 pKa = 10.04GVPQEE587 pKa = 4.15VMTPAPSNN595 pKa = 3.46
Molecular weight: 68.64 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6911 |
246 |
1235 |
767.9 |
88.0 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.73 ± 0.352 | 1.129 ± 0.191 |
5.643 ± 0.191 | 4.269 ± 0.201 |
3.921 ± 0.341 | 3.82 ± 0.296 |
3.241 ± 0.389 | 9.072 ± 0.169 |
3.617 ± 0.28 | 9.333 ± 0.343 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.243 ± 0.176 | 5.687 ± 0.394 |
5.296 ± 0.264 | 5.31 ± 0.15 |
5.498 ± 0.25 | 7.061 ± 0.262 |
8.494 ± 0.435 | 4.341 ± 0.183 |
0.868 ± 0.135 | 5.426 ± 0.381 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
